Árpi Vezér
Contrastive Mixture of Posteriors for Counterfactual Inference, Data Integration and Fairness
Jun 15, 2021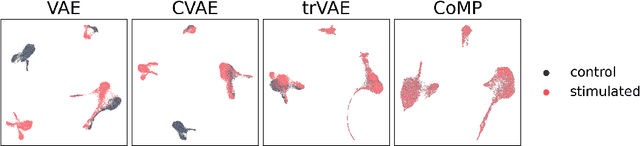
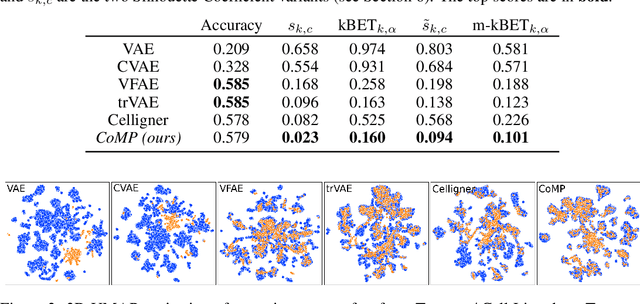
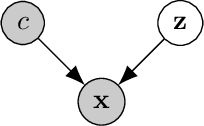
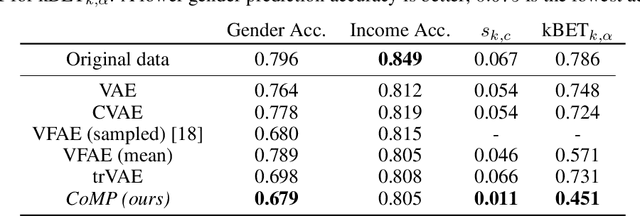
Abstract:Learning meaningful representations of data that can address challenges such as batch effect correction, data integration and counterfactual inference is a central problem in many domains including computational biology. Adopting a Conditional VAE framework, we identify the mathematical principle that unites these challenges: learning a representation that is marginally independent of a condition variable. We therefore propose the Contrastive Mixture of Posteriors (CoMP) method that uses a novel misalignment penalty to enforce this independence. This penalty is defined in terms of mixtures of the variational posteriors themselves, unlike prior work which uses external discrepancy measures such as MMD to ensure independence in latent space. We show that CoMP has attractive theoretical properties compared to previous approaches, especially when there is complex global structure in latent space. We further demonstrate state of the art performance on a number of real-world problems, including the challenging tasks of aligning human tumour samples with cancer cell-lines and performing counterfactual inference on single-cell RNA sequencing data. Incidentally, we find parallels with the fair representation learning literature, and demonstrate CoMP has competitive performance in learning fair yet expressive latent representations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge