Unsupervised single-particle deep clustering via statistical manifold learning
Paper and Code
Dec 31, 2016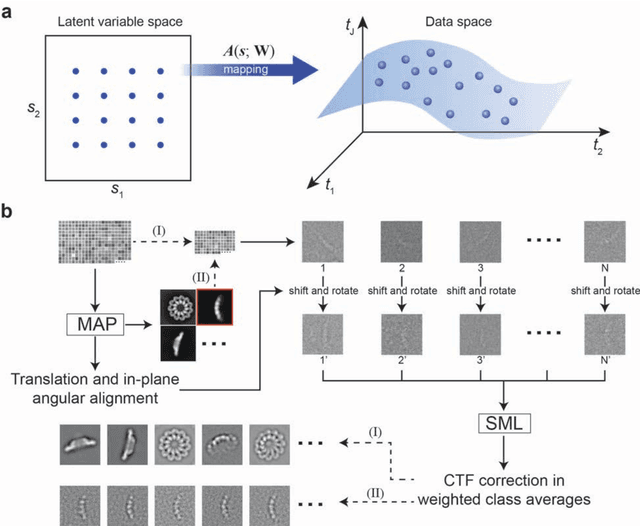
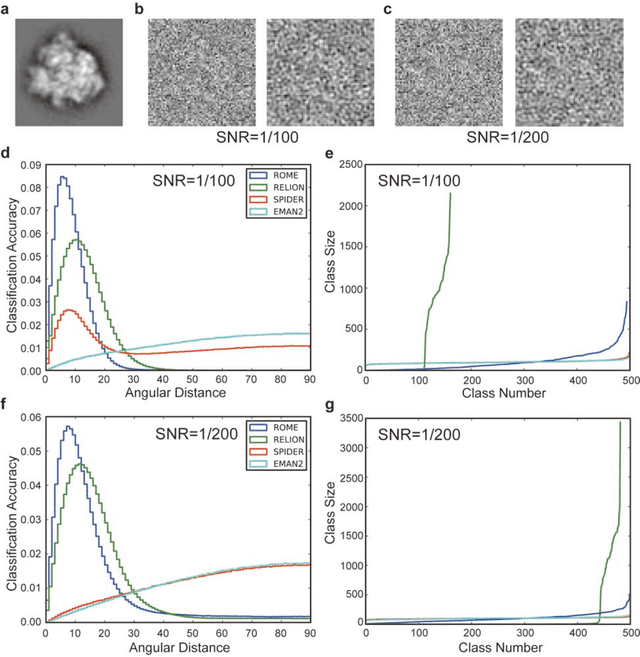
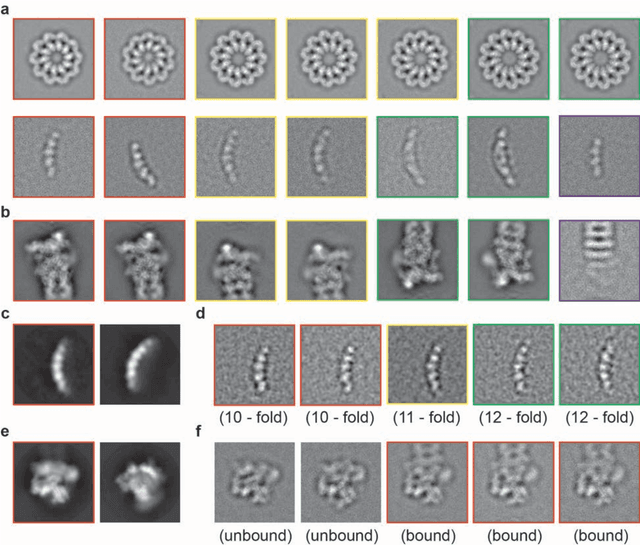
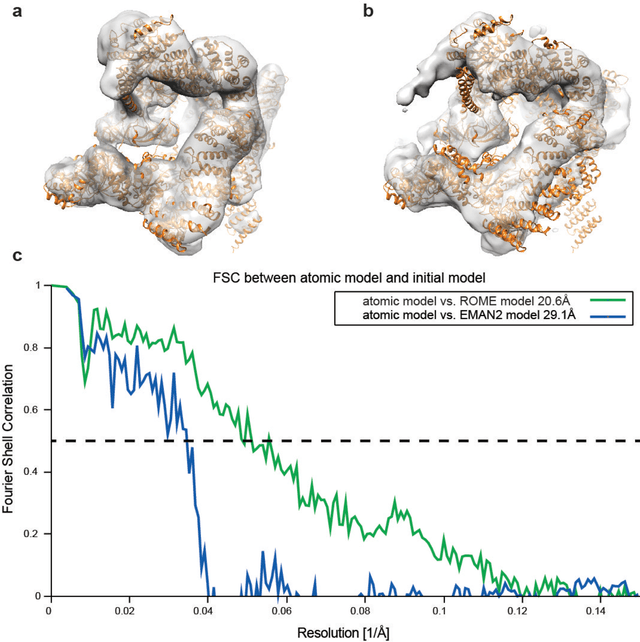
Motivation: Structural heterogeneity in single-particle cryo-electron microscopy (cryo-EM) data represents a major challenge for high-resolution structure determination. Unsupervised classification may serve as the first step in the assessment of structural heterogeneity. Traditional algorithms for unsupervised classification, such as K-means clustering and maximum likelihood optimization, may classify images into wrong classes with decreasing signal-to-noise-ratio (SNR) in the image data, yet demand increased cost in computation. Overcoming these limitations requires further development on clustering algorithms for high-performance cryo-EM data analysis. Results: Here we introduce a statistical manifold learning algorithm for unsupervised single-particle deep clustering. We show that statistical manifold learning improves classification accuracy by about 40% in the absence of input references for lower SNR data. Applications to several experimental datasets suggest that our deep clustering approach can detect subtle structural difference among classes. Through code optimization over the Intel high-performance computing (HPC) processors, our software implementation can generate thousands of reference-free class averages within several hours from hundreds of thousands of single-particle cryo-EM images, which allows significant improvement in ab initio 3D reconstruction resolution and quality. Our approach has been successfully applied in several structural determination projects. We expect that it provides a powerful computational tool in analyzing highly heterogeneous structural data and assisting in computational purification of single-particle datasets for high-resolution reconstruction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge