Unbiased and Efficient Log-Likelihood Estimation with Inverse Binomial Sampling
Paper and Code
Jan 12, 2020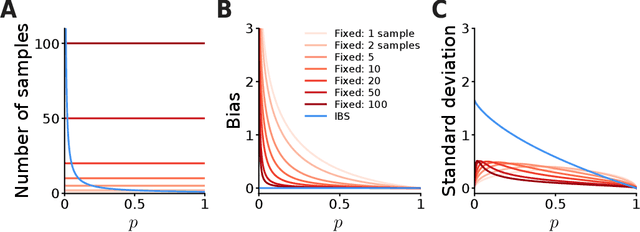
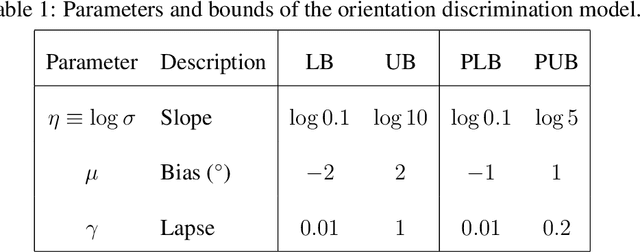
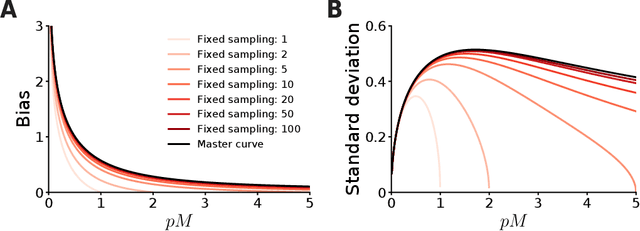

The fate of scientific hypotheses often relies on the ability of a computational model to explain the data, quantified in modern statistical approaches by the likelihood function. The log-likelihood is the key element for parameter estimation and model evaluation. However, the log-likelihood of complex models in fields such as computational biology and neuroscience is often intractable to compute analytically or numerically. In those cases, researchers can often only estimate the log-likelihood by comparing observed data with synthetic observations generated by model simulations. Standard techniques to approximate the likelihood via simulation either use summary statistics of the data or are at risk of producing severe biases in the estimate. Here, we explore another method, inverse binomial sampling (IBS), which can estimate the log-likelihood of an entire data set efficiently and without bias. For each observation, IBS draws samples from the simulator model until one matches the observation. The log-likelihood estimate is then a function of the number of samples drawn. The variance of this estimator is uniformly bounded, achieves the minimum variance for an unbiased estimator, and we can compute calibrated estimates of the variance. We provide theoretical arguments in favor of IBS and an empirical assessment of the method for maximum-likelihood estimation with simulation-based models. As case studies, we take three model-fitting problems of increasing complexity from computational and cognitive neuroscience. In all problems, IBS generally produces lower error in the estimated parameters and maximum log-likelihood values than alternative sampling methods with the same average number of samples. Our results demonstrate the potential of IBS as a practical, robust, and easy to implement method for log-likelihood evaluation when exact techniques are not available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge