TrialGraph: Machine Intelligence Enabled Insight from Graph Modelling of Clinical Trials
Paper and Code
Dec 15, 2021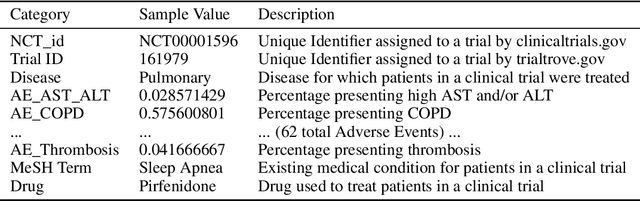
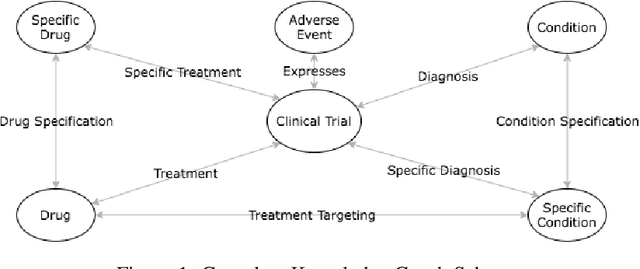
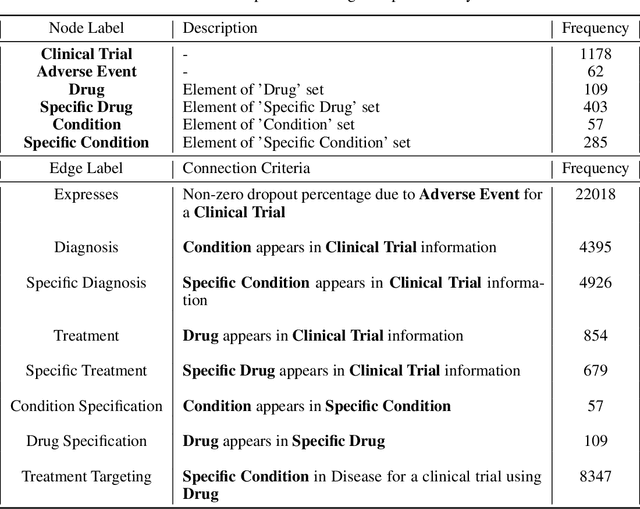

A major impediment to successful drug development is the complexity, cost, and scale of clinical trials. The detailed internal structure of clinical trial data can make conventional optimization difficult to achieve. Recent advances in machine learning, specifically graph-structured data analysis, have the potential to enable significant progress in improving the clinical trial design. TrialGraph seeks to apply these methodologies to produce a proof-of-concept framework for developing models which can aid drug development and benefit patients. In this work, we first introduce a curated clinical trial data set compiled from the CT.gov, AACT and TrialTrove databases (n=1191 trials; representing one million patients) and describe the conversion of this data to graph-structured formats. We then detail the mathematical basis and implementation of a selection of graph machine learning algorithms, which typically use standard machine classifiers on graph data embedded in a low-dimensional feature space. We trained these models to predict side effect information for a clinical trial given information on the disease, existing medical conditions, and treatment. The MetaPath2Vec algorithm performed exceptionally well, with standard Logistic Regression, Decision Tree, Random Forest, Support Vector, and Neural Network classifiers exhibiting typical ROC-AUC scores of 0.85, 0.68, 0.86, 0.80, and 0.77, respectively. Remarkably, the best performing classifiers could only produce typical ROC-AUC scores of 0.70 when trained on equivalent array-structured data. Our work demonstrates that graph modelling can significantly improve prediction accuracy on appropriate datasets. Successive versions of the project that refine modelling assumptions and incorporate more data types can produce excellent predictors with real-world applications in drug development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge