SUGAR: Subgraph Neural Network with Reinforcement Pooling and Self-Supervised Mutual Information Mechanism
Paper and Code
Jan 20, 2021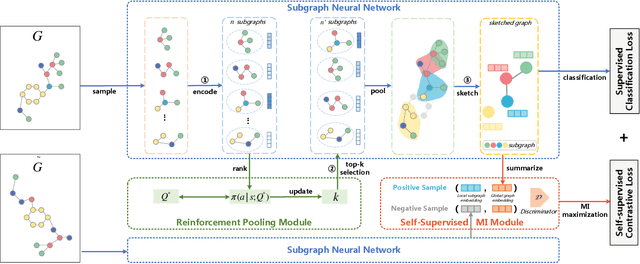
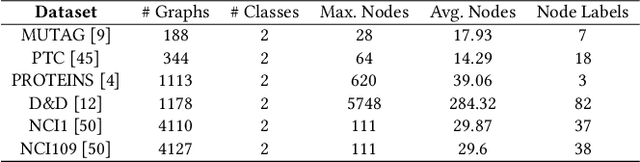
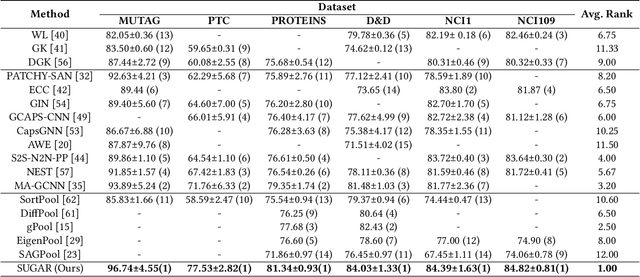
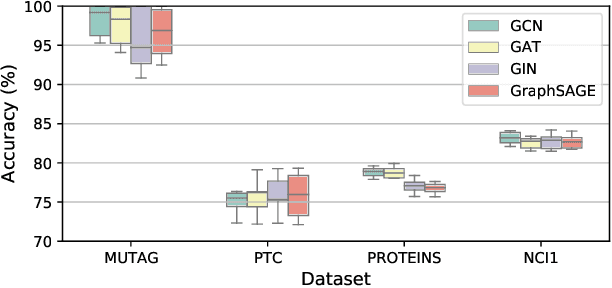
Graph representation learning has attracted increasing research attention. However, most existing studies fuse all structural features and node attributes to provide an overarching view of graphs, neglecting finer substructures' semantics, and suffering from interpretation enigmas. This paper presents a novel hierarchical subgraph-level selection and embedding based graph neural network for graph classification, namely SUGAR, to learn more discriminative subgraph representations and respond in an explanatory way. SUGAR reconstructs a sketched graph by extracting striking subgraphs as the representative part of the original graph to reveal subgraph-level patterns. To adaptively select striking subgraphs without prior knowledge, we develop a reinforcement pooling mechanism, which improves the generalization ability of the model. To differentiate subgraph representations among graphs, we present a self-supervised mutual information mechanism to encourage subgraph embedding to be mindful of the global graph structural properties by maximizing their mutual information. Extensive experiments on six typical bioinformatics datasets demonstrate a significant and consistent improvement in model quality with competitive performance and interpretability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge