Statistically Optimal Force Aggregation for Coarse-Graining Molecular Dynamics
Paper and Code
Feb 14, 2023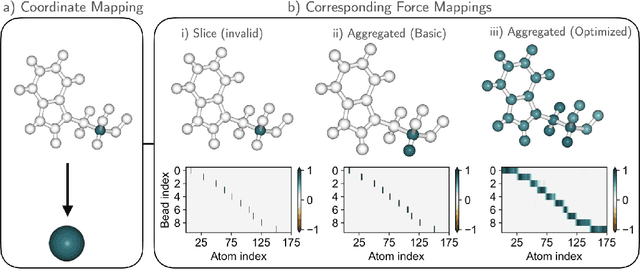
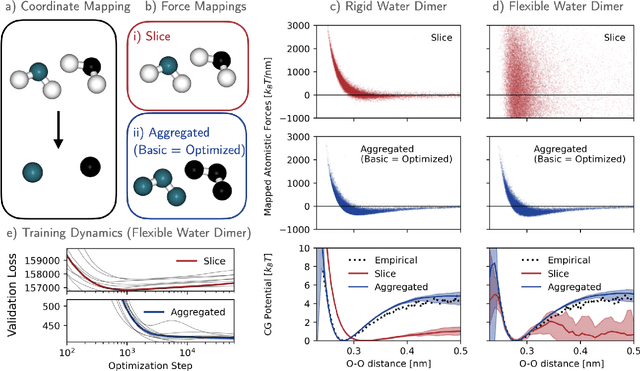
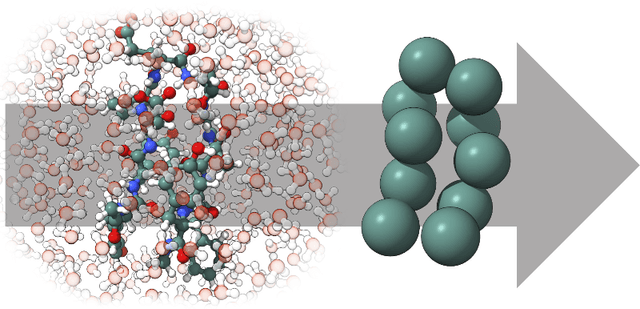
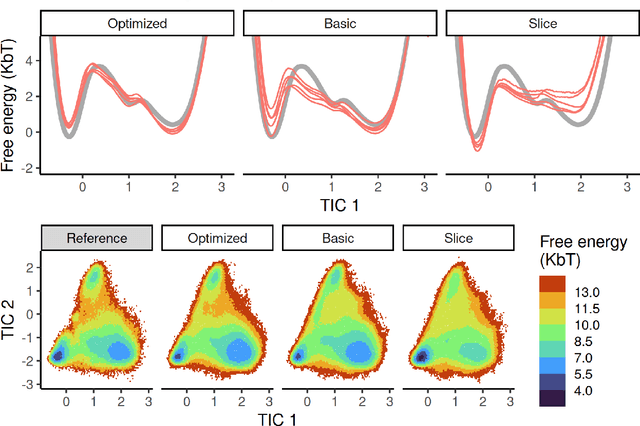
Machine-learned coarse-grained (CG) models have the potential for simulating large molecular complexes beyond what is possible with atomistic molecular dynamics. However, training accurate CG models remains a challenge. A widely used methodology for learning CG force-fields maps forces from all-atom molecular dynamics to the CG representation and matches them with a CG force-field on average. We show that there is flexibility in how to map all-atom forces to the CG representation, and that the most commonly used mapping methods are statistically inefficient and potentially even incorrect in the presence of constraints in the all-atom simulation. We define an optimization statement for force mappings and demonstrate that substantially improved CG force-fields can be learned from the same simulation data when using optimized force maps. The method is demonstrated on the miniproteins Chignolin and Tryptophan Cage and published as open-source code.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge