Stability selection enables robust learning of partial differential equations from limited noisy data
Paper and Code
Jul 17, 2019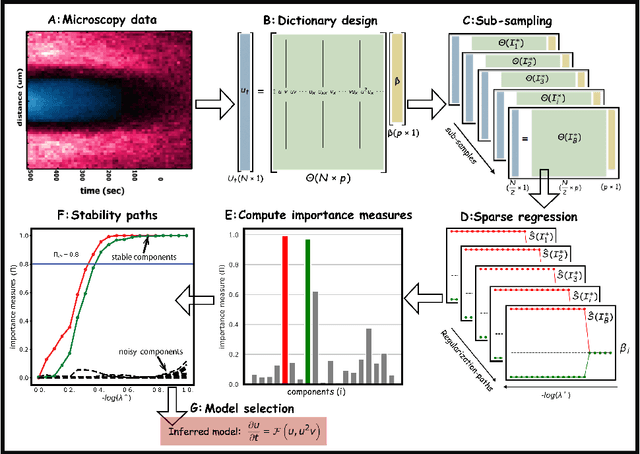
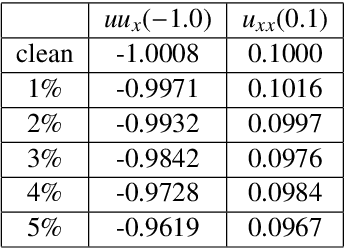
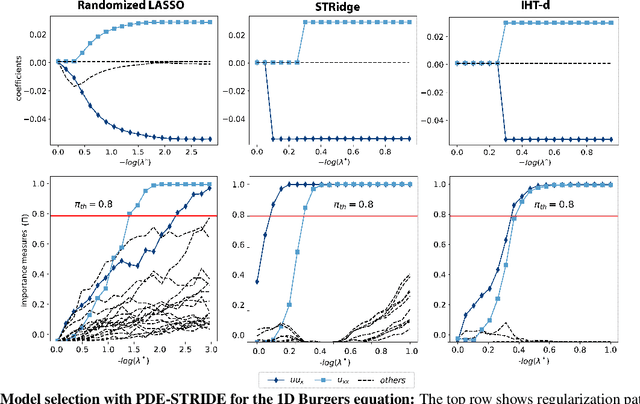

We present a statistical learning framework for robust identification of partial differential equations from noisy spatiotemporal data. Extending previous sparse regression approaches for inferring PDE models from simulated data, we address key issues that have thus far limited the application of these methods to noisy experimental data, namely their robustness against noise and the need for manual parameter tuning. We address both points by proposing a stability-based model selection scheme to determine the level of regularization required for reproducible recovery of the underlying PDE. This avoids manual parameter tuning and provides a principled way to improve the method's robustness against noise in the data. Our stability selection approach, termed PDE-STRIDE, can be combined with any sparsity-promoting penalized regression model and provides an interpretable criterion for model component importance. We show that in particular the combination of stability selection with the iterative hard-thresholding algorithm from compressed sensing provides a fast, parameter-free, and robust computational framework for PDE inference that outperforms previous algorithmic approaches with respect to recovery accuracy, amount of data required, and robustness to noise. We illustrate the performance of our approach on a wide range of noise-corrupted simulated benchmark problems, including 1D Burgers, 2D vorticity-transport, and 3D reaction-diffusion problems. We demonstrate the practical applicability of our method on real-world data by considering a purely data-driven re-evaluation of the advective triggering hypothesis for an embryonic polarization system in C.~elegans. Using fluorescence microscopy images of C.~elegans zygotes as input data, our framework is able to recover the PDE model for the regulatory reaction-diffusion-flow network of the associated proteins.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge