SMILES2Vec: An Interpretable General-Purpose Deep Neural Network for Predicting Chemical Properties
Paper and Code
Mar 18, 2018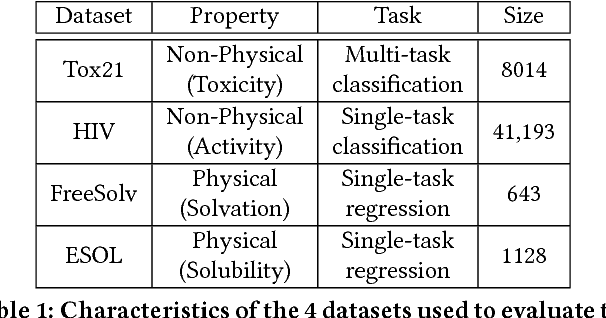
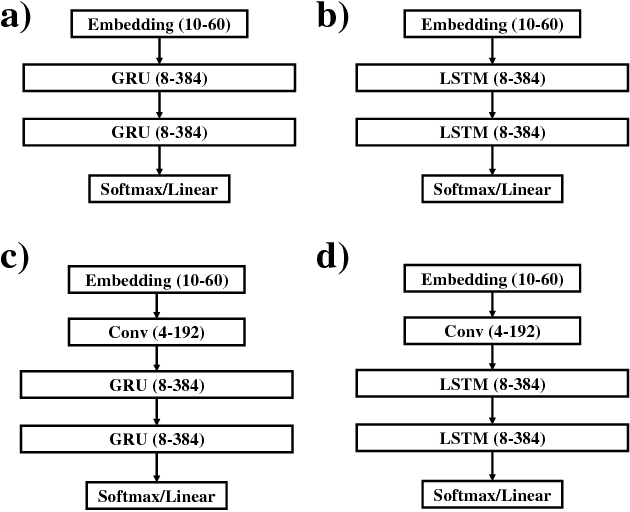
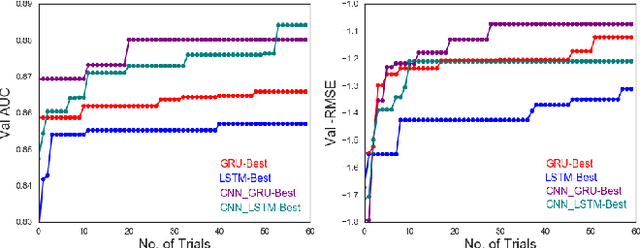
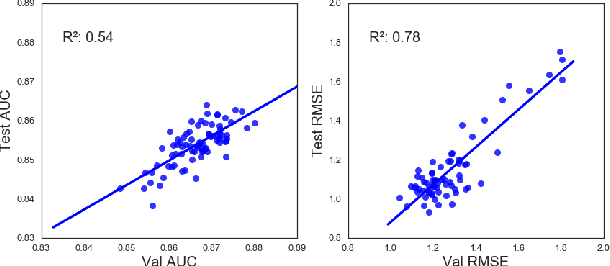
Chemical databases store information in text representations, and the SMILES format is a universal standard used in many cheminformatics software. Encoded in each SMILES string is structural information that can be used to predict complex chemical properties. In this work, we develop SMILES2vec, a deep RNN that automatically learns features from SMILES to predict chemical properties, without the need for additional explicit feature engineering. Using Bayesian optimization methods to tune the network architecture, we show that an optimized SMILES2vec model can serve as a general-purpose neural network for predicting distinct chemical properties including toxicity, activity, solubility and solvation energy, while also outperforming contemporary MLP neural networks that uses engineered features. Furthermore, we demonstrate proof-of-concept of interpretability by developing an explanation mask that localizes on the most important characters used in making a prediction. When tested on the solubility dataset, it identified specific parts of a chemical that is consistent with established first-principles knowledge with an accuracy of 88%. Our work demonstrates that neural networks can learn technically accurate chemical concept and provide state-of-the-art accuracy, making interpretable deep neural networks a useful tool of relevance to the chemical industry.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge