Semi-supervised, Topology-Aware Segmentation of Tubular Structures from Live Imaging 3D Microscopy
Paper and Code
May 20, 2021
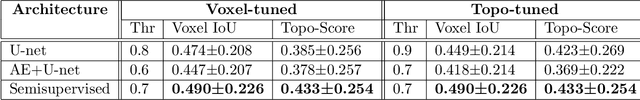


Motivated by a challenging tubular network segmentation task, this paper tackles two commonly encountered problems in biomedical imaging: Topological consistency of the segmentation, and limited annotations. We propose a topological score which measures both topological and geometric consistency between the predicted and ground truth segmentations, applied for model selection and validation. We apply our topological score in three scenarios: i. a U-net ii. a U-net pretrained on an autoencoder, and iii. a semisupervised U-net architecture, which offers a straightforward approach to jointly training the network both as an autoencoder and a segmentation algorithm. This allows us to utilize un-annotated data for training a representation that generalizes across test data variability, in spite of our annotated training data having very limited variation. Our contributions are validated on a challenging segmentation task, locating tubular structures in the fetal pancreas from noisy live imaging confocal microscopy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge