Self-supervised Representation Learning of Neuronal Morphologies
Paper and Code
Jan 18, 2022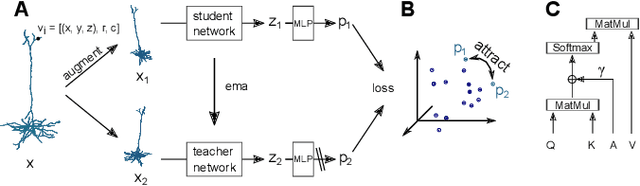
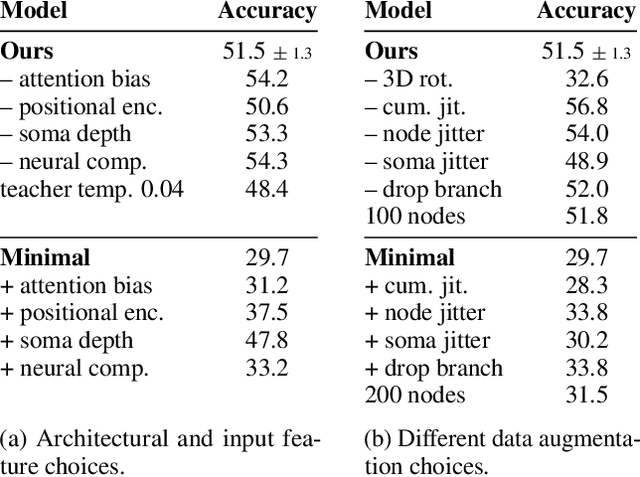
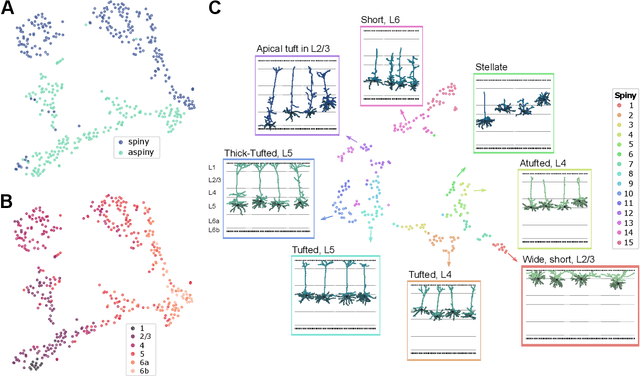
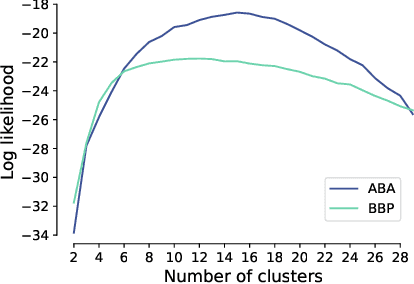
Understanding the diversity of cell types and their function in the brain is one of the key challenges in neuroscience. The advent of large-scale datasets has given rise to the need of unbiased and quantitative approaches to cell type classification. We present GraphDINO, a purely data-driven approach to learning a low dimensional representation of the 3D morphology of neurons. GraphDINO is a novel graph representation learning method for spatial graphs utilizing self-supervised learning on transformer models. It combines attention-based global interaction between nodes and classic graph convolutional processing. We show, in two different species and cortical areas, that this method is able to yield morphological cell type clustering that is comparable to manual feature-based classification and shows a good correspondence to expert-labeled cell types. Our method is applicable beyond neuroscience in settings where samples in a dataset are graphs and graph-level embeddings are desired.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge