Segmentation of Defective Skulls from CT Data for Tissue Modelling
Paper and Code
Nov 20, 2019
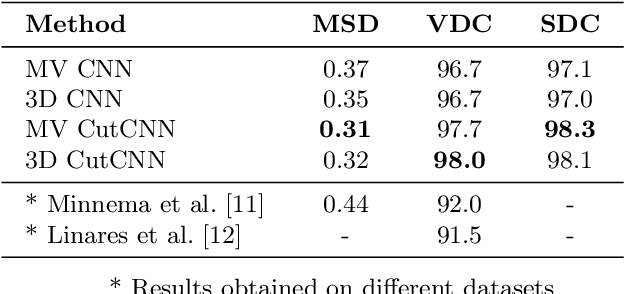


In this work we present a method of automatic segmentation of defective skulls for custom cranial implant design and 3D printing purposes. Since such tissue models are usually required in patient cases with complex anatomical defects and variety of external objects present in the acquired data, most deep learning-based approaches fall short because it is not possible to create a sufficient training dataset that would encompass the spectrum of all possible structures. Because CNN segmentation experiments in this application domain have been so far limited to simple patch-based CNN architectures, we first show how the usage of the encoder-decoder architecture can substantially improve the segmentation accuracy. Then, we show how the number of segmentation artifacts, which usually require manual corrections, can be further reduced by adding a boundary term to CNN training and by globally optimizing the segmentation with graph-cut. Finally, we show that using the proposed method, 3D segmentation accurate enough for clinical application can be achieved with 2D CNN architectures as well as their 3D counterparts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge