RPA: Probabilistic analysis of probe performance and robust summarization
Paper and Code
Apr 06, 2013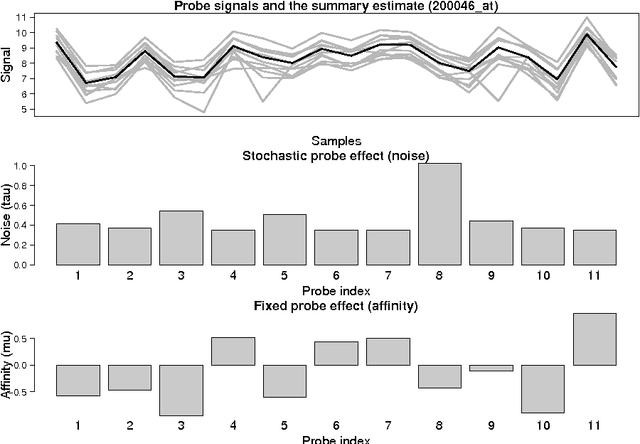
Probe-level models have led to improved performance in microarray studies but the various sources of probe-level contamination are still poorly understood. Data-driven analysis of probe performance can be used to quantify the uncertainty in individual probes and to highlight the relative contribution of different noise sources. Improved understanding of the probe-level effects can lead to improved preprocessing techniques and microarray design. We have implemented probabilistic tools for probe performance analysis and summarization on short oligonucleotide arrays. In contrast to standard preprocessing approaches, the methods provide quantitative estimates of probe-specific noise and affinity terms and tools to investigate these parameters. Tools to incorporate prior information of the probes in the analysis are provided as well. Comparisons to known probe-level error sources and spike-in data sets validate the approach. Implementation is freely available in R/BioConductor: http://www.bioconductor.org/packages/release/bioc/html/RPA.html
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge