Quantifying the effect of image compression on supervised learning applications in optical microscopy
Paper and Code
Sep 26, 2020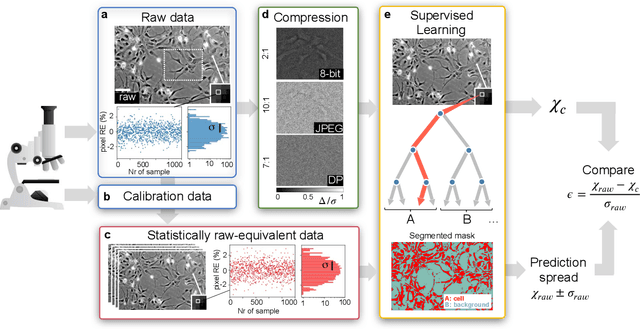
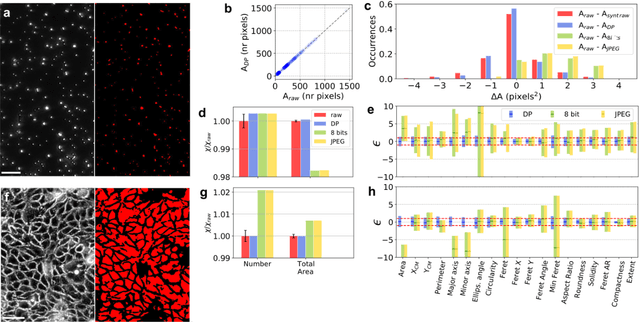
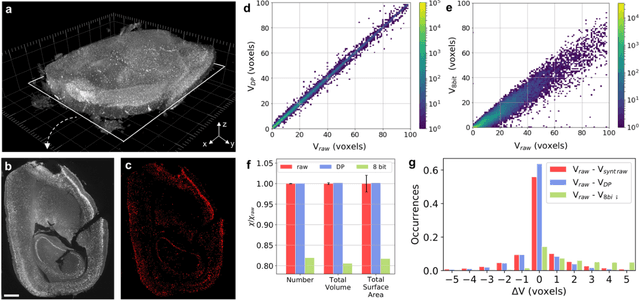
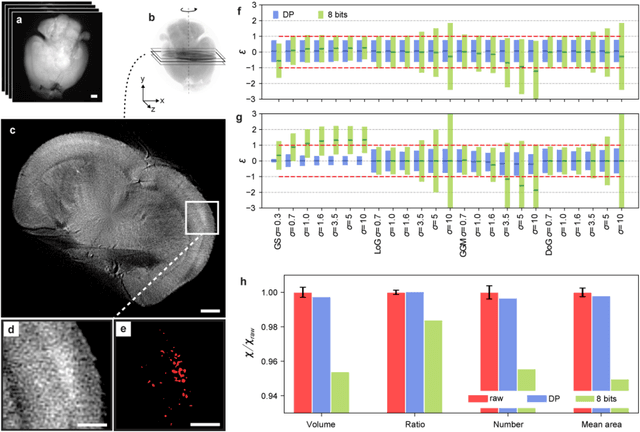
The impressive growth of data throughput in optical microscopy has triggered a widespread use of supervised learning (SL) models running on compressed image datasets for efficient automated analysis. However, since lossy image compression risks to produce unpredictable artifacts, quantifying the effect of data compression on SL applications is of pivotal importance to assess their reliability, especially for clinical use. We propose an experimental method to evaluate the tolerability of image compression distortions in 2D and 3D cell segmentation SL tasks: predictions on compressed data are compared to the raw predictive uncertainty, which is numerically estimated from the raw noise statistics measured through sensor calibration. We show that predictions on object- and image-specific segmentation parameters can be altered by up to 15% and more than 10 standard deviations after 16-to-8 bits downsampling or JPEG compression. In contrast, a recently developed lossless compression algorithm provides a prediction spread which is statistically equivalent to that stemming from raw noise, while providing a compression ratio of up to 10:1. By setting a lower bound to the SL predictive uncertainty, our technique can be generalized to validate a variety of data analysis pipelines in SL-assisted fields.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge