Principal component-guided sparse regression
Paper and Code
Oct 24, 2018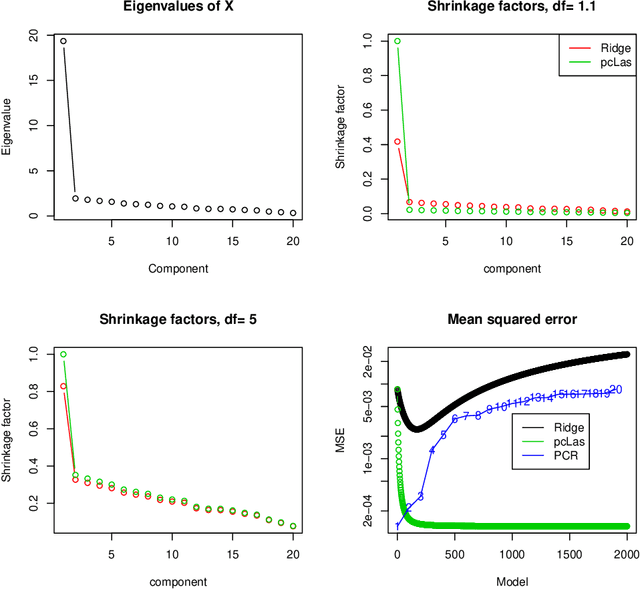
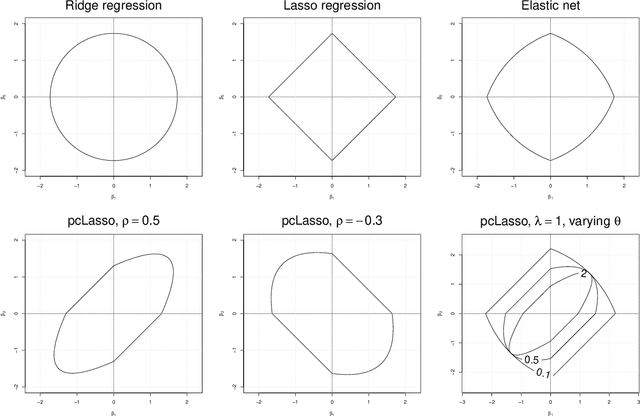

We propose a new method for supervised learning, especially suited to wide data where the number of features is much greater than the number of observations. The method combines the lasso ($\ell_1$) sparsity penalty with a quadratic penalty that shrinks the coefficient vector toward the leading principal components of the feature matrix. We call the proposed method the "principal components lasso" ("pcLasso"). The method can be especially powerful if the features are pre-assigned to groups (such as cell-pathways, assays or protein interaction networks). In that case, pcLasso shrinks each group-wise component of the solution toward the leading principal components of that group. In the process, it also carries out selection of the feature groups. We provide some theory for this method and illustrate it on a number of simulated and real data examples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge