Predicting microsatellite instability and key biomarkers in colorectal cancer from H&E-stained images: Achieving SOTA with Less Data using Swin Transformer
Paper and Code
Aug 22, 2022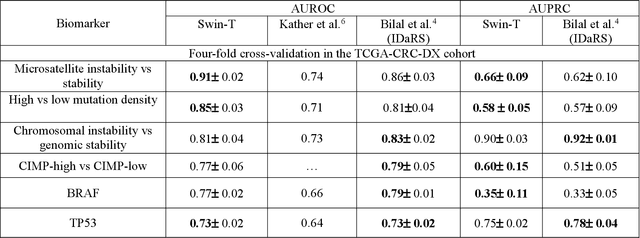
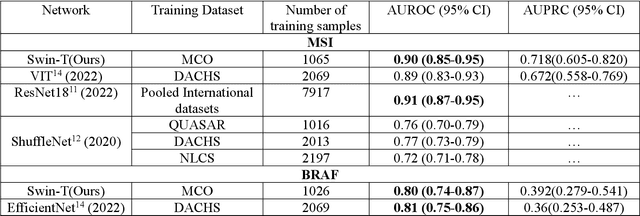
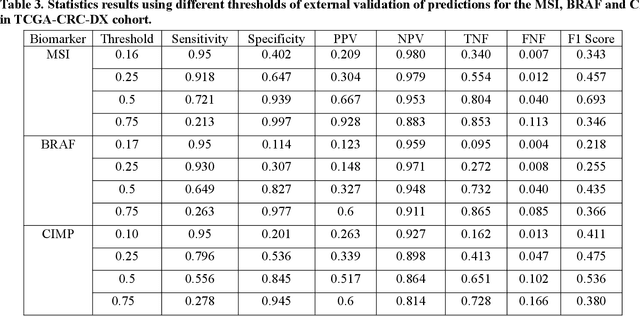
Artificial intelligence (AI) models have been developed for predicting clinically relevant biomarkers, including microsatellite instability (MSI), for colorectal cancers (CRC). However, the current deep-learning networks are data-hungry and require large training datasets, which are often lacking in the medical domain. In this study, based on the latest Hierarchical Vision Transformer using Shifted Windows (Swin-T), we developed an efficient workflow for biomarkers in CRC (MSI, hypermutation, chromosomal instability, CpG island methylator phenotype, BRAF, and TP53 mutation) that only required relatively small datasets, but achieved the state-of-the-art (SOTA) predictive performance. Our Swin-T workflow not only substantially outperformed published models in an intra-study cross-validation experiment using TCGA-CRC-DX dataset (N = 462), but also showed excellent generalizability in cross-study external validation and delivered a SOTA AUROC of 0.90 for MSI using the MCO dataset for training (N = 1065) and the same TCGA-CRC-DX for testing. Similar performance (AUROC=0.91) was achieved by Echle and colleagues using 8000 training samples (ResNet18) on the same testing dataset. Swin-T was extremely efficient using small training datasets and exhibits robust predictive performance with only 200-500 training samples. These data indicate that Swin-T may be 5-10 times more efficient than the current state-of-the-art algorithms for MSI based on ResNet18 and ShuffleNet. Furthermore, the Swin-T models showed promise as pre-screening tests for MSI status and BRAF mutation status, which could exclude and reduce the samples before the subsequent standard testing in a cascading diagnostic workflow to allow turnaround time reduction and cost saving.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge