PotatoGANs: Utilizing Generative Adversarial Networks, Instance Segmentation, and Explainable AI for Enhanced Potato Disease Identification and Classification
Paper and Code
May 12, 2024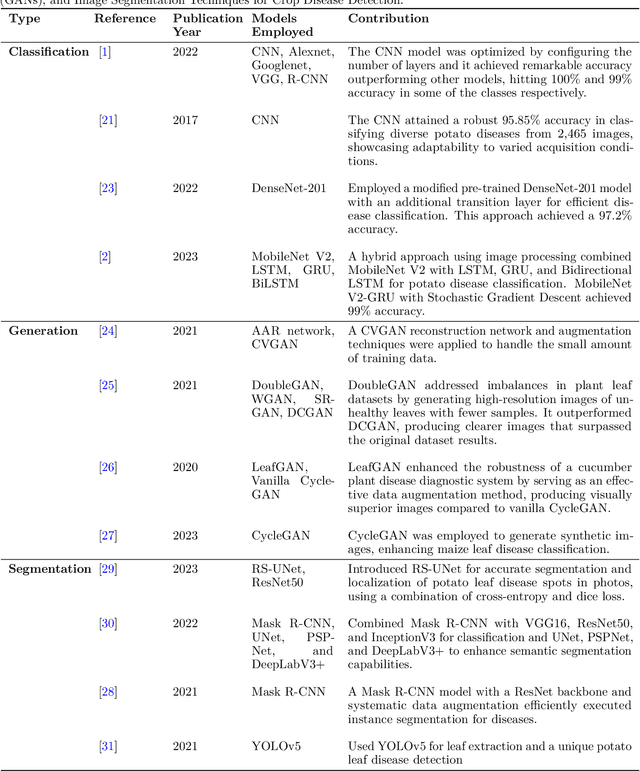
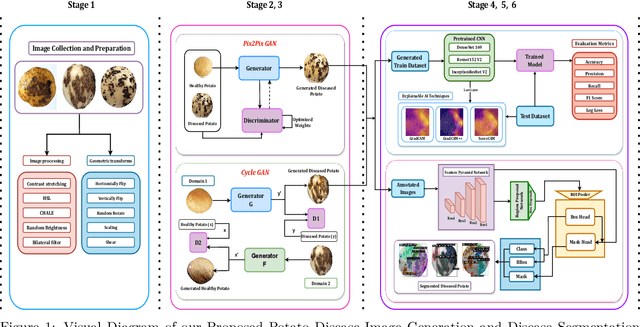
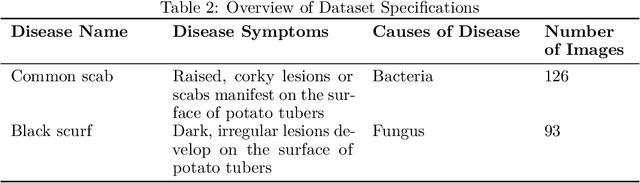
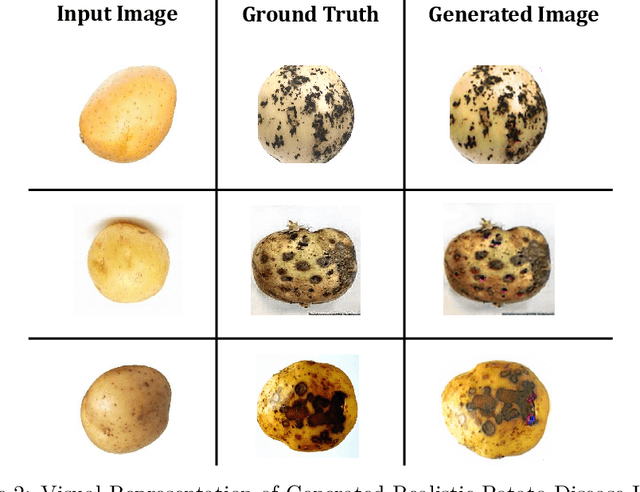
Numerous applications have resulted from the automation of agricultural disease segmentation using deep learning techniques. However, when applied to new conditions, these applications frequently face the difficulty of overfitting, resulting in lower segmentation performance. In the context of potato farming, where diseases have a large influence on yields, it is critical for the agricultural economy to quickly and properly identify these diseases. Traditional data augmentation approaches, such as rotation, flip, and translation, have limitations and frequently fail to provide strong generalization results. To address these issues, our research employs a novel approach termed as PotatoGANs. In this novel data augmentation approach, two types of Generative Adversarial Networks (GANs) are utilized to generate synthetic potato disease images from healthy potato images. This approach not only expands the dataset but also adds variety, which helps to enhance model generalization. Using the Inception score as a measure, our experiments show the better quality and realisticness of the images created by PotatoGANs, emphasizing their capacity to resemble real disease images closely. The CycleGAN model outperforms the Pix2Pix GAN model in terms of image quality, as evidenced by its higher IS scores CycleGAN achieves higher Inception scores (IS) of 1.2001 and 1.0900 for black scurf and common scab, respectively. This synthetic data can significantly improve the training of large neural networks. It also reduces data collection costs while enhancing data diversity and generalization capabilities. Our work improves interpretability by combining three gradient-based Explainable AI algorithms (GradCAM, GradCAM++, and ScoreCAM) with three distinct CNN architectures (DenseNet169, Resnet152 V2, InceptionResNet V2) for potato disease classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge