Path-Augmented Graph Transformer Network
Paper and Code
May 29, 2019
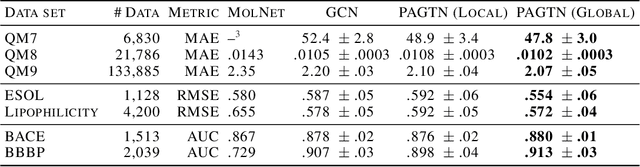


Much of the recent work on learning molecular representations has been based on Graph Convolution Networks (GCN). These models rely on local aggregation operations and can therefore miss higher-order graph properties. To remedy this, we propose Path-Augmented Graph Transformer Networks (PAGTN) that are explicitly built on longer-range dependencies in graph-structured data. Specifically, we use path features in molecular graphs to create global attention layers. We compare our PAGTN model against the GCN model and show that our model consistently outperforms GCNs on molecular property prediction datasets including quantum chemistry (QM7, QM8, QM9), physical chemistry (ESOL, Lipophilictiy) and biochemistry (BACE, BBBP).
* Appears in ICML LRG Workshop
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge