Non-Volatile Memory Accelerated Geometric Multi-Scale Resolution Analysis
Paper and Code
Feb 21, 2022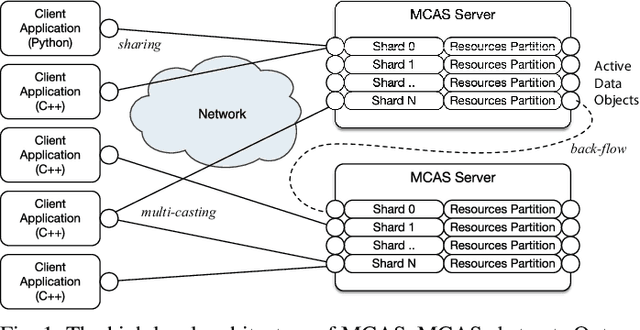
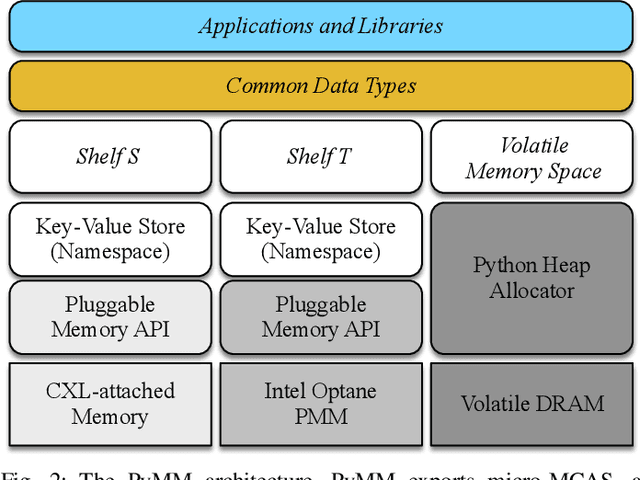
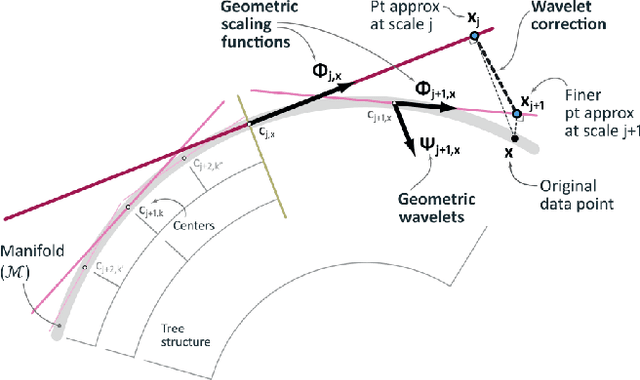
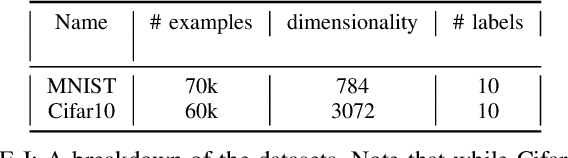
Dimensionality reduction algorithms are standard tools in a researcher's toolbox. Dimensionality reduction algorithms are frequently used to augment downstream tasks such as machine learning, data science, and also are exploratory methods for understanding complex phenomena. For instance, dimensionality reduction is commonly used in Biology as well as Neuroscience to understand data collected from biological subjects. However, dimensionality reduction techniques are limited by the von-Neumann architectures that they execute on. Specifically, data intensive algorithms such as dimensionality reduction techniques often require fast, high capacity, persistent memory which historically hardware has been unable to provide at the same time. In this paper, we present a re-implementation of an existing dimensionality reduction technique called Geometric Multi-Scale Resolution Analysis (GMRA) which has been accelerated via novel persistent memory technology called Memory Centric Active Storage (MCAS). Our implementation uses a specialized version of MCAS called PyMM that provides native support for Python datatypes including NumPy arrays and PyTorch tensors. We compare our PyMM implementation against a DRAM implementation, and show that when data fits in DRAM, PyMM offers competitive runtimes. When data does not fit in DRAM, our PyMM implementation is still able to process the data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge