Non-negative Subspace Feature Representation for Few-shot Learning in Medical Imaging
Paper and Code
Apr 04, 2024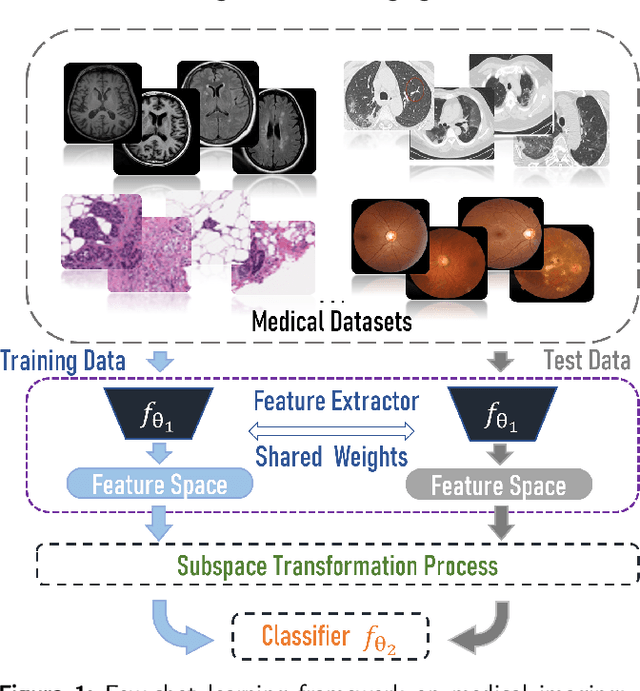
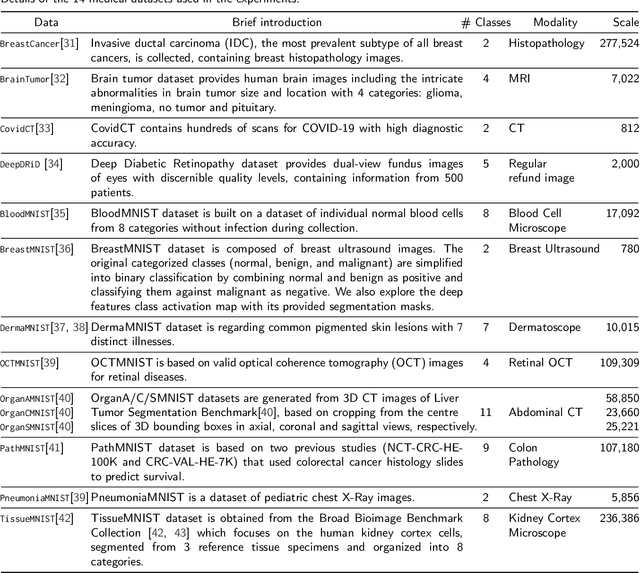
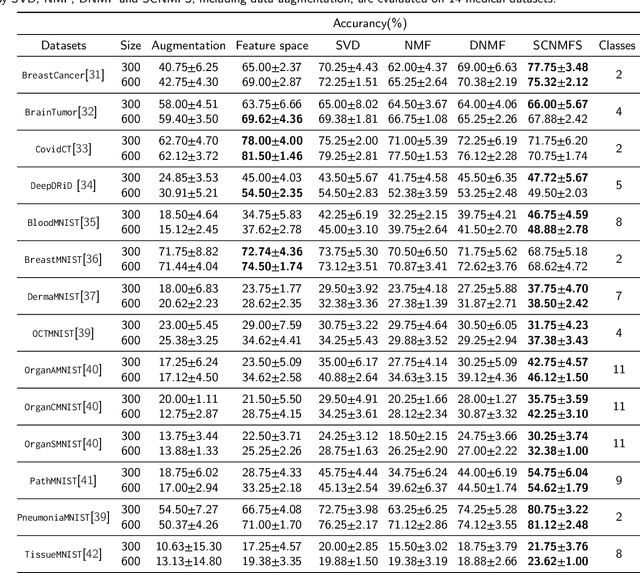
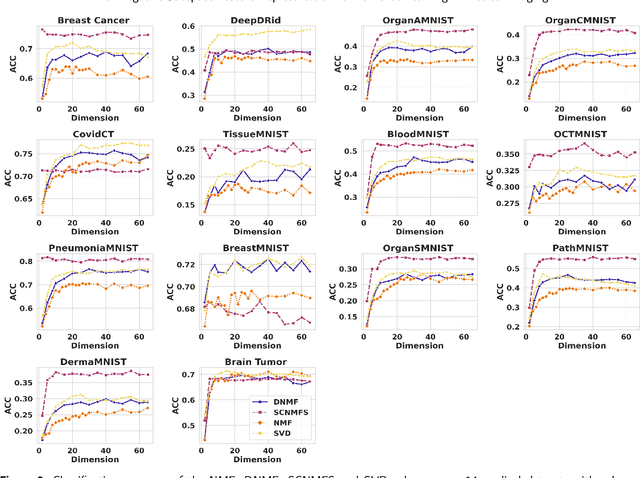
Unlike typical visual scene recognition domains, in which massive datasets are accessible to deep neural networks, medical image interpretations are often obstructed by the paucity of data. In this paper, we investigate the effectiveness of data-based few-shot learning in medical imaging by exploring different data attribute representations in a low-dimensional space. We introduce different types of non-negative matrix factorization (NMF) in few-shot learning, addressing the data scarcity issue in medical image classification. Extensive empirical studies are conducted in terms of validating the effectiveness of NMF, especially its supervised variants (e.g., discriminative NMF, and supervised and constrained NMF with sparseness), and the comparison with principal component analysis (PCA), i.e., the collaborative representation-based dimensionality reduction technique derived from eigenvectors. With 14 different datasets covering 11 distinct illness categories, thorough experimental results and comparison with related techniques demonstrate that NMF is a competitive alternative to PCA for few-shot learning in medical imaging, and the supervised NMF algorithms are more discriminative in the subspace with greater effectiveness. Furthermore, we show that the part-based representation of NMF, especially its supervised variants, is dramatically impactful in detecting lesion areas in medical imaging with limited samples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge