Moment Matching Deep Contrastive Latent Variable Models
Paper and Code
Feb 21, 2022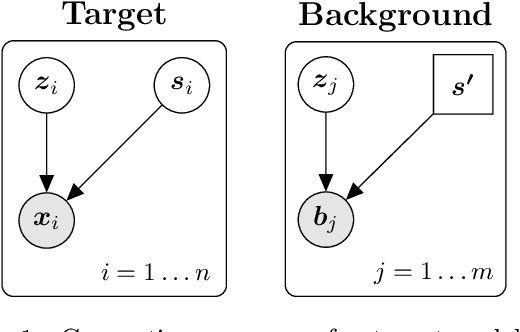


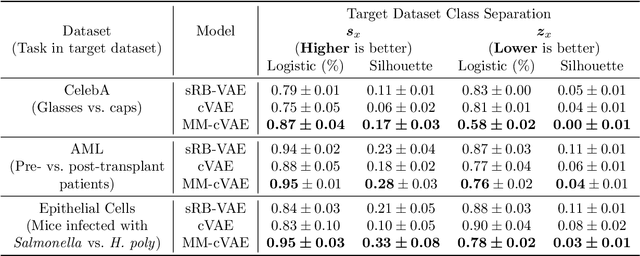
In the contrastive analysis (CA) setting, machine learning practitioners are specifically interested in discovering patterns that are enriched in a target dataset as compared to a background dataset generated from sources of variation irrelevant to the task at hand. For example, a biomedical data analyst may seek to understand variations in genomic data only present among patients with a given disease as opposed to those also present in healthy control subjects. Such scenarios have motivated the development of contrastive latent variable models to isolate variations unique to these target datasets from those shared across the target and background datasets, with current state of the art models based on the variational autoencoder (VAE) framework. However, previously proposed models do not explicitly enforce the constraints on latent variables underlying CA, potentially leading to the undesirable leakage of information between the two sets of latent variables. Here we propose the moment matching contrastive VAE (MM-cVAE), a reformulation of the VAE for CA that uses the maximum mean discrepancy to explicitly enforce two crucial latent variable constraints underlying CA. On three challenging CA tasks we find that our method outperforms the previous state-of-the-art both qualitatively and on a set of quantitative metrics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge