Molecular modeling with machine-learned universal potential functions
Paper and Code
Mar 06, 2021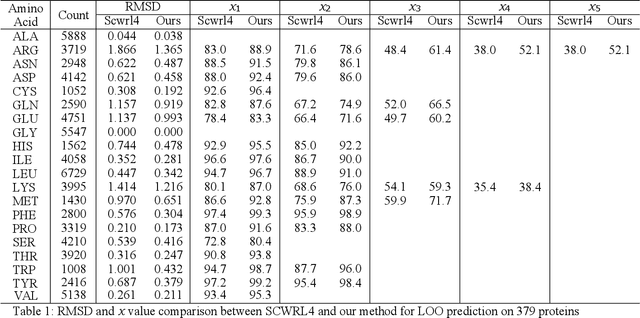
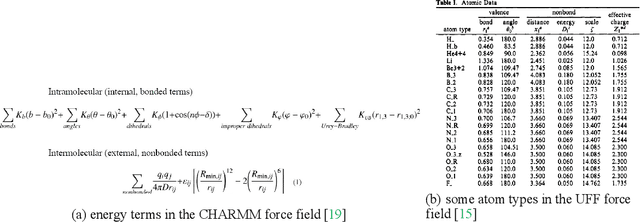
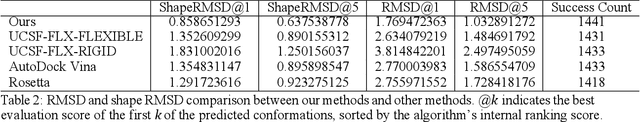
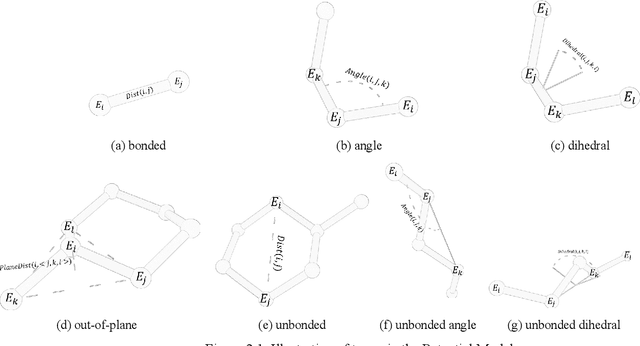
Molecular modeling is an important topic in drug discovery. Decades of research have led to the development of high quality scalable molecular force fields. In this paper, we show that neural networks can be used to train an universal approximator for energy potential functions. By incorporating a fully automated training process we have been able to train smooth, differentiable, and predictive potential functions on large scale crystal structures. A variety of tests have also performed to show the superiority and versatility of the machine-learned model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge