JANUS: Parallel Tempered Genetic Algorithm Guided by Deep Neural Networks for Inverse Molecular Design
Paper and Code
Jun 07, 2021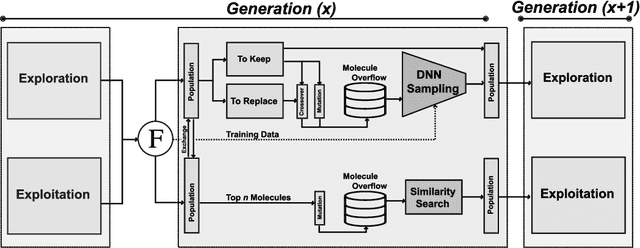
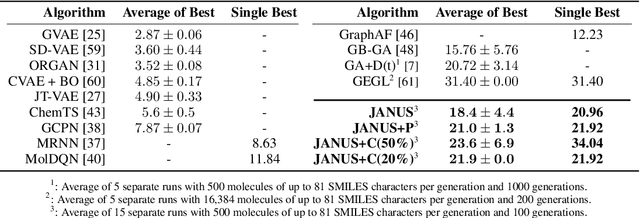
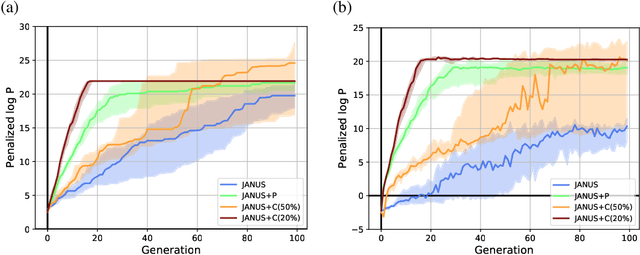
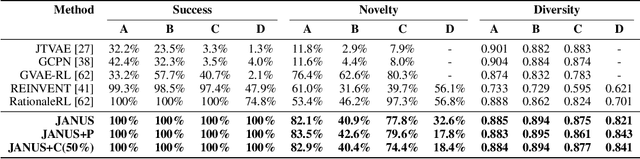
Inverse molecular design, i.e., designing molecules with specific target properties, can be posed as an optimization problem. High-dimensional optimization tasks in the natural sciences are commonly tackled via population-based metaheuristic optimization algorithms such as evolutionary algorithms. However, expensive property evaluation, which is often required, can limit the widespread use of such approaches as the associated cost can become prohibitive. Herein, we present JANUS, a genetic algorithm that is inspired by parallel tempering. It propagates two populations, one for exploration and another for exploitation, improving optimization by reducing expensive property evaluations. Additionally, JANUS is augmented by a deep neural network that approximates molecular properties via active learning for enhanced sampling of the chemical space. Our method uses the SELFIES molecular representation and the STONED algorithm for the efficient generation of structures, and outperforms other generative models in common inverse molecular design tasks achieving state-of-the-art performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge