IP-UNet: Intensity Projection UNet Architecture for 3D Medical Volume Segmentation
Paper and Code
Aug 24, 2023

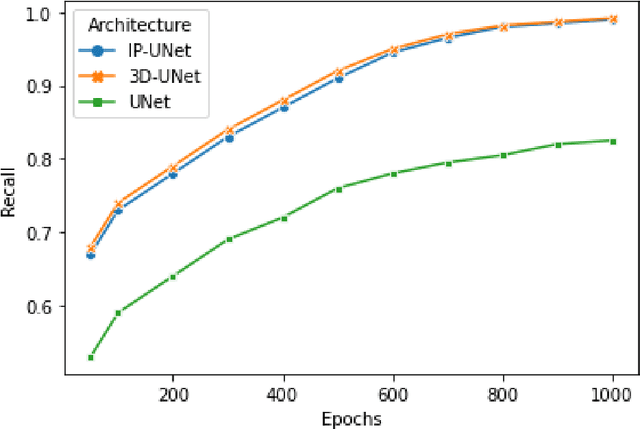
CNNs have been widely applied for medical image analysis. However, limited memory capacity is one of the most common drawbacks of processing high-resolution 3D volumetric data. 3D volumes are usually cropped or downsized first before processing, which can result in a loss of resolution, increase class imbalance, and affect the performance of the segmentation algorithms. In this paper, we propose an end-to-end deep learning approach called IP-UNet. IP-UNet is a UNet-based model that performs multi-class segmentation on Intensity Projection (IP) of 3D volumetric data instead of the memory-consuming 3D volumes. IP-UNet uses limited memory capability for training without losing the original 3D image resolution. We compare the performance of three models in terms of segmentation accuracy and computational cost: 1) Slice-by-slice 2D segmentation of the CT scan images using a conventional 2D UNet model. 2) IP-UNet that operates on data obtained by merging the extracted Maximum Intensity Projection (MIP), Closest Vessel Projection (CVP), and Average Intensity Projection (AvgIP) representations of the source 3D volumes, then applying the UNet model on the output IP images. 3) 3D-UNet model directly reads the 3D volumes constructed from a series of CT scan images and outputs the 3D volume of the predicted segmentation. We test the performance of these methods on 3D volumetric images for automatic breast calcification detection. Experimental results show that IP-Unet can achieve similar segmentation accuracy with 3D-Unet but with much better performance. It reduces the training time by 70\% and memory consumption by 92\%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge