Interpreting Temporal Graph Neural Networks with Koopman Theory
Paper and Code
Oct 17, 2024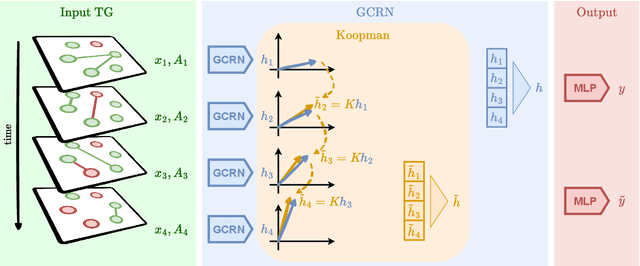
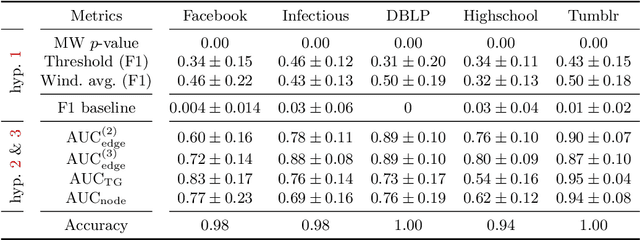
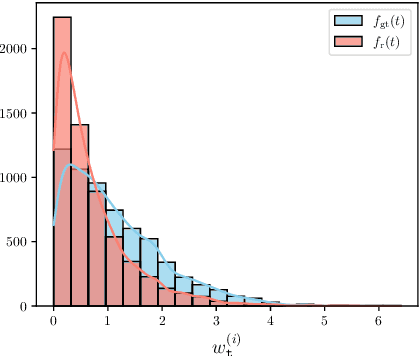
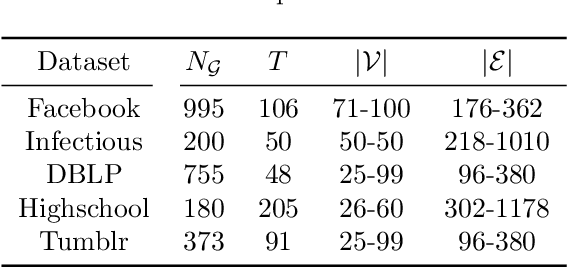
Spatiotemporal graph neural networks (STGNNs) have shown promising results in many domains, from forecasting to epidemiology. However, understanding the dynamics learned by these models and explaining their behaviour is significantly more complex than for models dealing with static data. Inspired by Koopman theory, which allows a simpler description of intricate, nonlinear dynamical systems, we introduce an explainability approach for temporal graphs. We present two methods to interpret the STGNN's decision process and identify the most relevant spatial and temporal patterns in the input for the task at hand. The first relies on dynamic mode decomposition (DMD), a Koopman-inspired dimensionality reduction method. The second relies on sparse identification of nonlinear dynamics (SINDy), a popular method for discovering governing equations, which we use for the first time as a general tool for explainability. We show how our methods can correctly identify interpretable features such as infection times and infected nodes in the context of dissemination processes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge