Interpretable Graph Convolutional Neural Networks for Inference on Noisy Knowledge Graphs
Paper and Code
Dec 01, 2018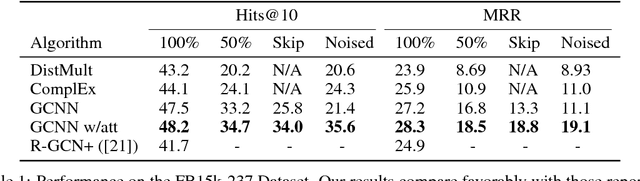
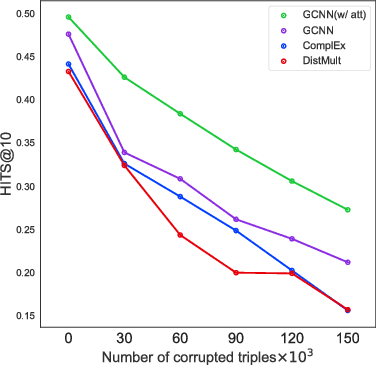
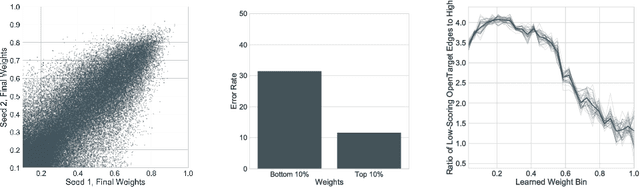
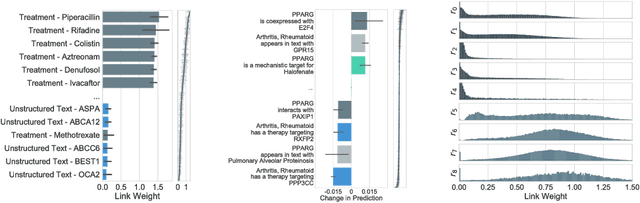
In this work, we provide a new formulation for Graph Convolutional Neural Networks (GCNNs) for link prediction on graph data that addresses common challenges for biomedical knowledge graphs (KGs). We introduce a regularized attention mechanism to GCNNs that not only improves performance on clean datasets, but also favorably accommodates noise in KGs, a pervasive issue in real-world applications. Further, we explore new visualization methods for interpretable modelling and to illustrate how the learned representation can be exploited to automate dataset denoising. The results are demonstrated on a synthetic dataset, the common benchmark dataset FB15k-237, and a large biomedical knowledge graph derived from a combination of noisy and clean data sources. Using these improvements, we visualize a learned model's representation of the disease cystic fibrosis and demonstrate how to interrogate a neural network to show the potential of PPARG as a candidate therapeutic target for rheumatoid arthritis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge