IFSS-Net: Interactive Few-Shot Siamese Network for Faster Muscles Segmentation and Propagation in 3-D Freehand Ultrasound
Paper and Code
Nov 26, 2020
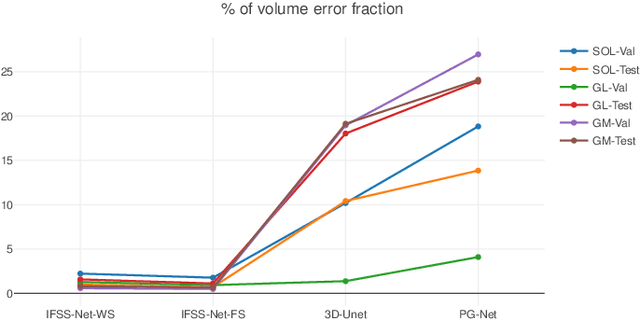


We present an accurate, fast and efficient method for segmentation and muscle mask propagation in 3D freehand ultrasound data, towards accurate volume quantification. To this end, we propose a deep Siamese 3D Encoder-Decoder network that captures the evolution of the muscle appearance and shape for contiguous slices and uses it to propagate a reference mask annotated by a clinical expert. To handle longer changes of the muscle shape over the entire volume and to provide an accurate propagation, we devised a Bidirectional Long Short Term Memory module. To train our model with a minimal amount of training samples, we propose a strategy to combine learning from few annotated 2D ultrasound slices with sequential pseudo-labeling of the unannotated slices. To promote few-shot learning, we propose a decremental update of the objective function to guide the model convergence in the absence of large amounts of annotated data. Finally, to handle the class-imbalance between foreground and background muscle pixels, we propose a parametric Tversky loss function that learns to adaptively penalize false positives and false negatives. We validate our approach for the segmentation, label propagation, and volume computation of the three low-limb muscles on a dataset of 44 subjects. We achieve a dice score coefficient of over $95~\%$ and a small fraction of error with $1.6035~\pm~0.587$.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge