Identification of Effective Connectivity Subregions
Paper and Code
Aug 08, 2019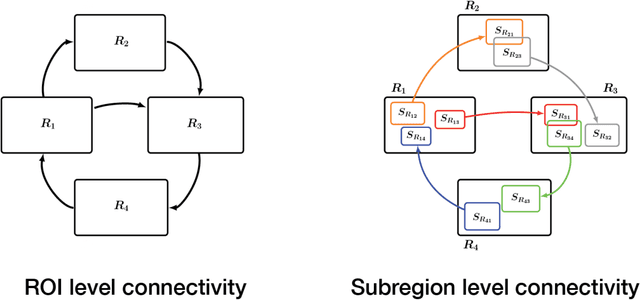
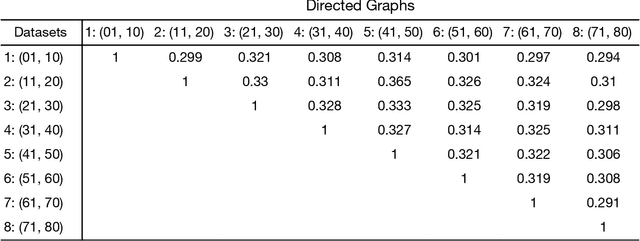
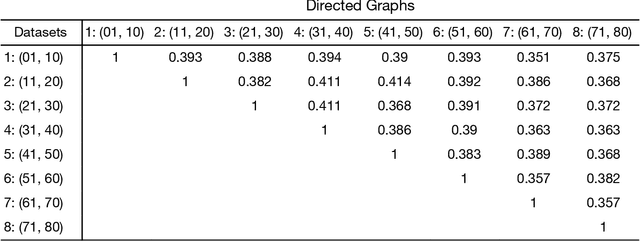
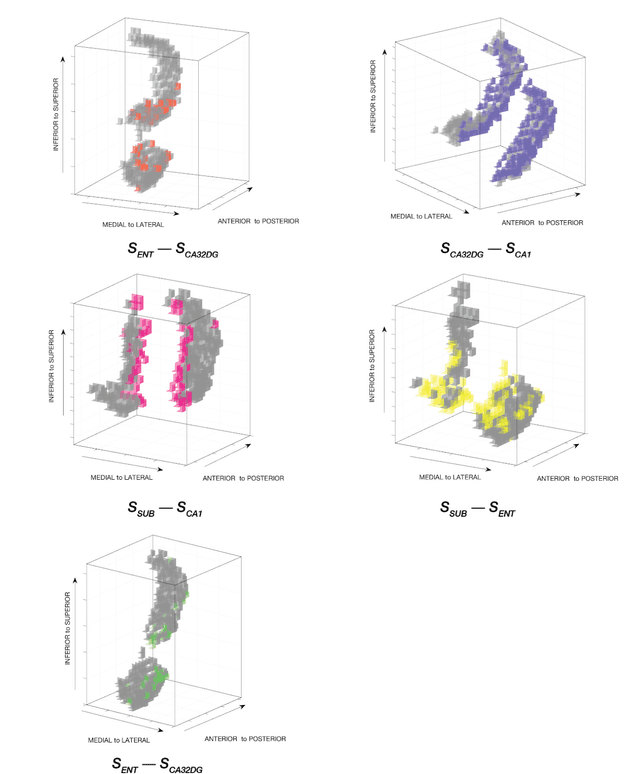
Standard fMRI connectivity analyses depend on aggregating the time series of individual voxels within regions of interest (ROIs). In certain cases, this spatial aggregation implies a loss of valuable functional and anatomical information about smaller subsets of voxels that drive the ROI level connectivity. We use two recently published graphical search methods to identify subsets of voxels that are highly responsible for the connectivity between larger ROIs. To illustrate the procedure, we apply both methods to longitudinal high-resolution resting state fMRI data from regions in the medial temporal lobe from a single individual. Both methods recovered similar subsets of voxels within larger ROIs of entorhinal cortex and hippocampus subfields that also show spatial consistency across different scanning sessions and across hemispheres. In contrast to standard functional connectivity methods, both algorithms applied here are robust against false positive connections produced by common causes and indirect paths (in contrast to Pearson's correlation) and common effect conditioning (in contrast to partial correlation based approaches). These algorithms allow for identification of subregions of voxels driving the connectivity between regions of interest, recovering valuable anatomical and functional information that is lost when ROIs are aggregated. Both methods are specially suited for voxelwise connectivity research, given their running times and scalability to big data problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge