Highdicom: A Python library for standardized encoding of image annotations and machine learning model outputs in pathology and radiology
Paper and Code
Jun 14, 2021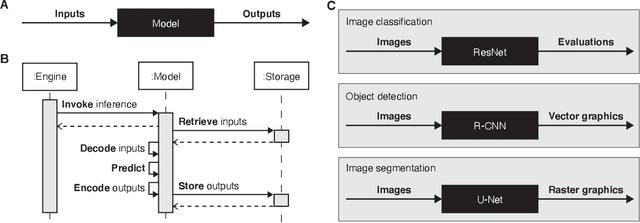
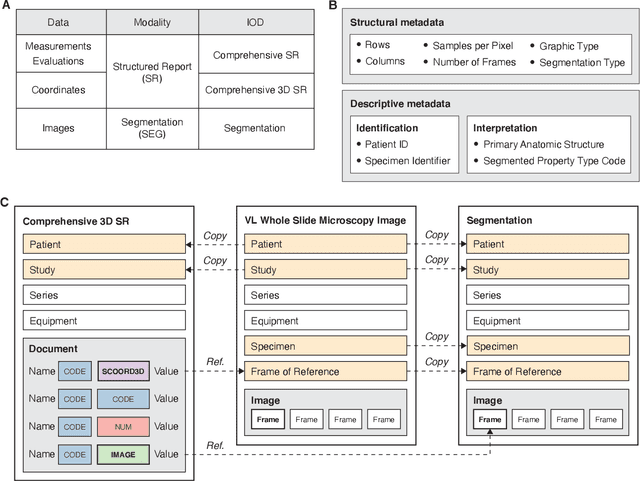
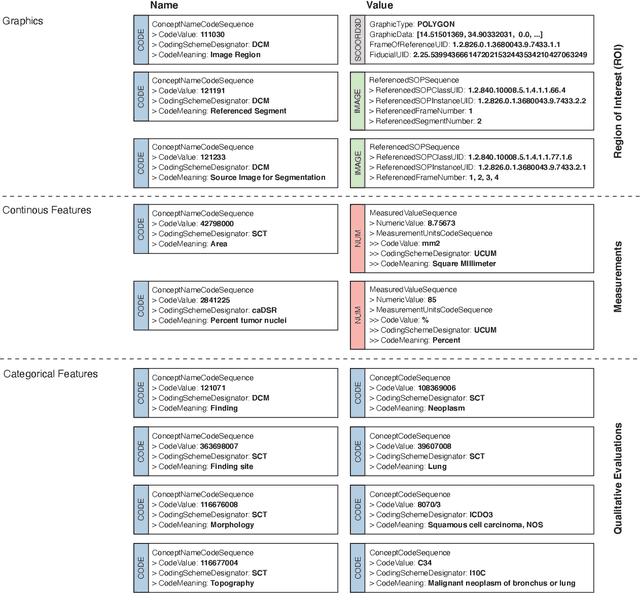
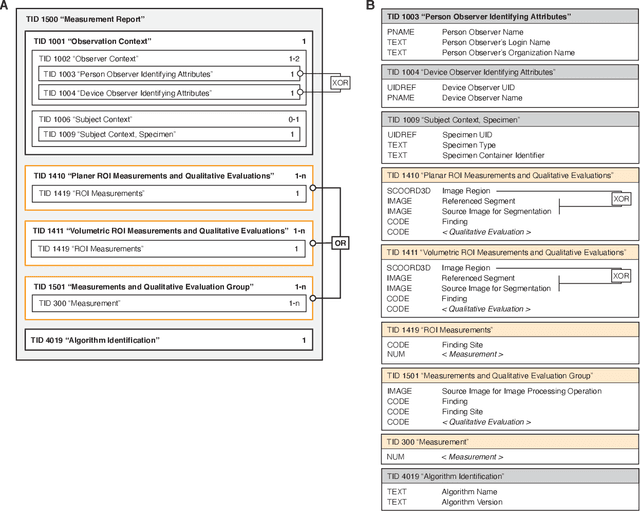
Machine learning is revolutionizing image-based diagnostics in pathology and radiology. ML models have shown promising results in research settings, but their lack of interoperability has been a major barrier for clinical integration and evaluation. The DICOM a standard specifies Information Object Definitions and Services for the representation and communication of digital images and related information, including image-derived annotations and analysis results. However, the complexity of the standard represents an obstacle for its adoption in the ML community and creates a need for software libraries and tools that simplify working with data sets in DICOM format. Here we present the highdicom library, which provides a high-level application programming interface for the Python programming language that abstracts low-level details of the standard and enables encoding and decoding of image-derived information in DICOM format in a few lines of Python code. The highdicom library ties into the extensive Python ecosystem for image processing and machine learning. Simultaneously, by simplifying creation and parsing of DICOM-compliant files, highdicom achieves interoperability with the medical imaging systems that hold the data used to train and run ML models, and ultimately communicate and store model outputs for clinical use. We demonstrate through experiments with slide microscopy and computed tomography imaging, that, by bridging these two ecosystems, highdicom enables developers to train and evaluate state-of-the-art ML models in pathology and radiology while remaining compliant with the DICOM standard and interoperable with clinical systems at all stages. To promote standardization of ML research and streamline the ML model development and deployment process, we made the library available free and open-source.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge