Harnessing Simulation for Molecular Embeddings
Paper and Code
Feb 04, 2023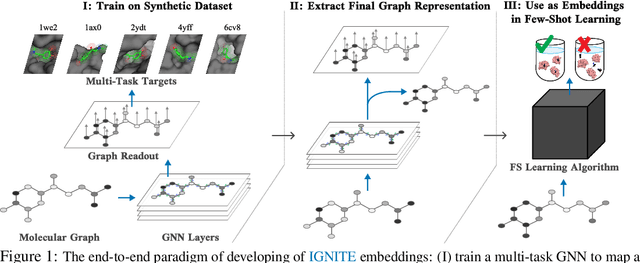
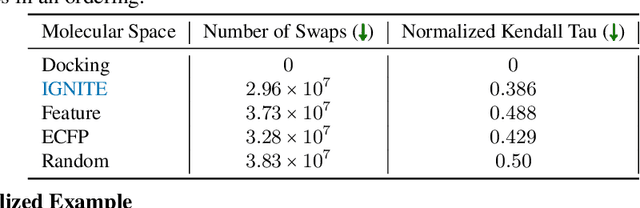
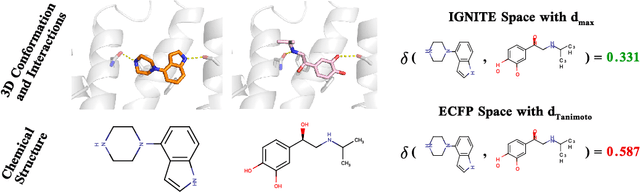
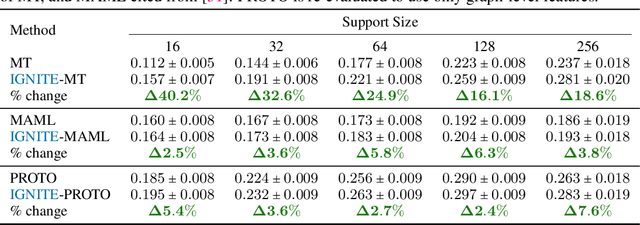
While deep learning has unlocked advances in computational biology once thought to be decades away, extending deep learning techniques to the molecular domain has proven challenging, as labeled data is scarce and the benefit from self-supervised learning can be negligible in many cases. In this work, we explore a different approach. Inspired by methods in deep reinforcement learning and robotics, we explore harnessing physics-based molecular simulation to develop molecular embeddings. By fitting a Graph Neural Network to simulation data, molecules that display similar interactions with biological targets under simulation develop similar representations in the embedding space. These embeddings can then be used to initialize the feature space of down-stream models trained on real-world data to encode information learned during simulation into a molecular prediction task. Our experimental findings indicate this approach improves the performance of existing deep learning models on real-world molecular prediction tasks by as much as 38% with minimal modification to the downstream model and no hyperparameter tuning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge