GIT-Mol: A Multi-modal Large Language Model for Molecular Science with Graph, Image, and Text
Paper and Code
Aug 14, 2023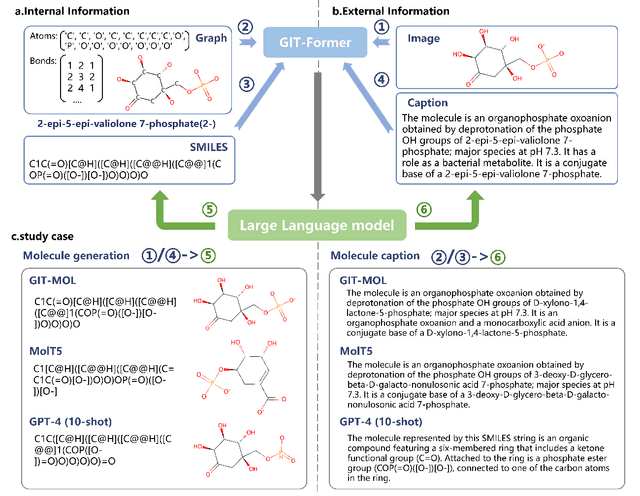

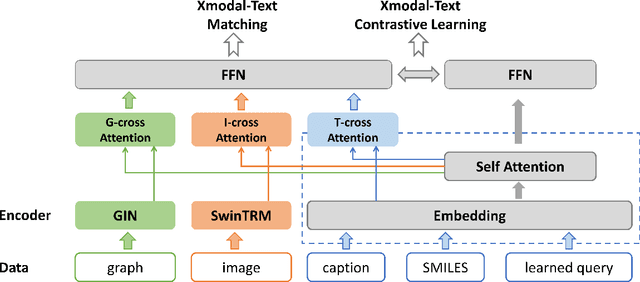
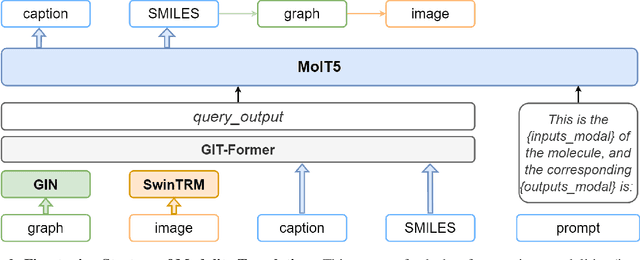
Large language models have made significant strides in natural language processing, paving the way for innovative applications including molecular representation and generation. However, most existing single-modality approaches cannot capture the abundant and complex information in molecular data. Here, we introduce GIT-Mol, a multi-modal large language model that integrates the structure Graph, Image, and Text information, including the Simplified Molecular Input Line Entry System (SMILES) and molecular captions. To facilitate the integration of multi-modal molecular data, we propose GIT-Former, a novel architecture capable of mapping all modalities into a unified latent space. Our study develops an innovative any-to-language molecular translation strategy and achieves a 10%-15% improvement in molecular captioning, a 5%-10% accuracy increase in property prediction, and a 20% boost in molecule generation validity compared to baseline or single-modality models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge