Feature Selection in the Contrastive Analysis Setting
Paper and Code
Oct 27, 2023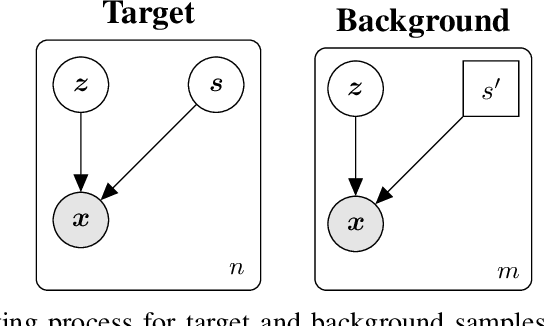
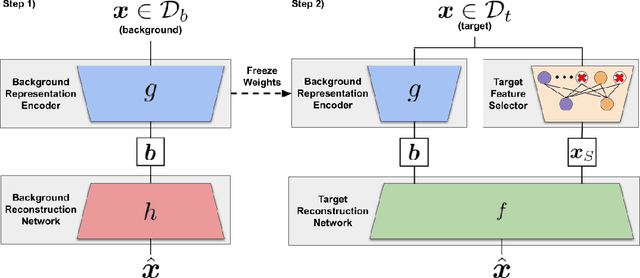
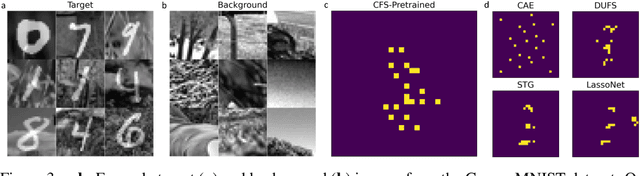

Contrastive analysis (CA) refers to the exploration of variations uniquely enriched in a target dataset as compared to a corresponding background dataset generated from sources of variation that are irrelevant to a given task. For example, a biomedical data analyst may wish to find a small set of genes to use as a proxy for variations in genomic data only present among patients with a given disease (target) as opposed to healthy control subjects (background). However, as of yet the problem of feature selection in the CA setting has received little attention from the machine learning community. In this work we present contrastive feature selection (CFS), a method for performing feature selection in the CA setting. We motivate our approach with a novel information-theoretic analysis of representation learning in the CA setting, and we empirically validate CFS on a semi-synthetic dataset and four real-world biomedical datasets. We find that our method consistently outperforms previously proposed state-of-the-art supervised and fully unsupervised feature selection methods not designed for the CA setting. An open-source implementation of our method is available at https://github.com/suinleelab/CFS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge