Extreme Consistency: Overcoming Annotation Scarcity and Domain Shifts
Paper and Code
Apr 15, 2020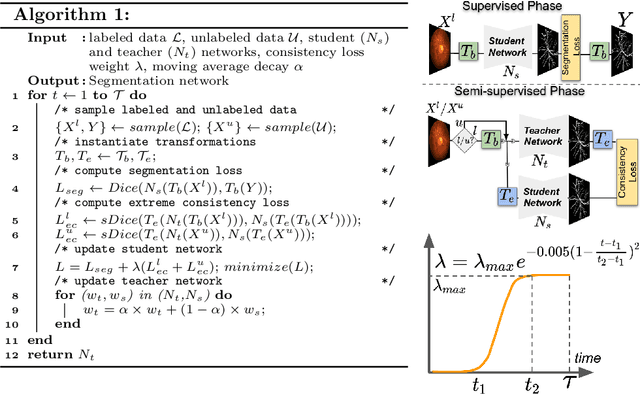
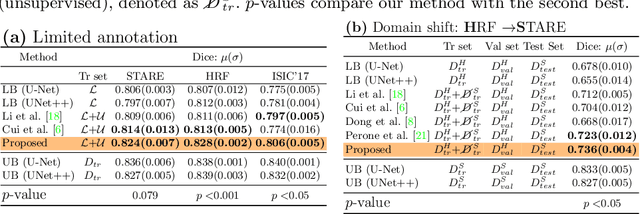
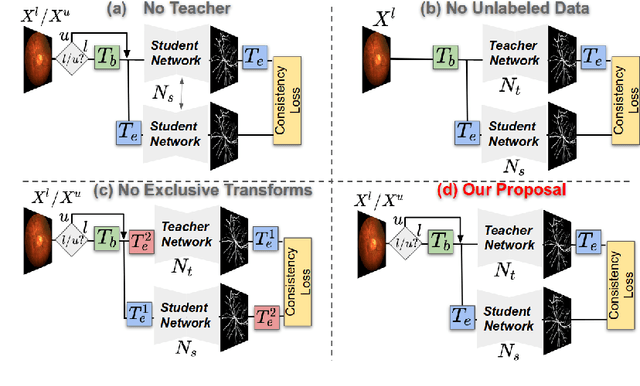
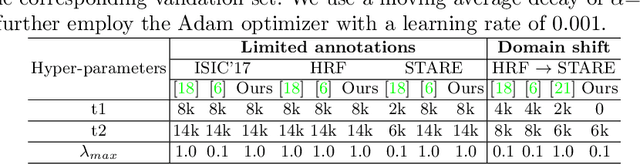
Supervised learning has proved effective for medical image analysis. However, it can utilize only the small labeled portion of data; it fails to leverage the large amounts of unlabeled data that is often available in medical image datasets. Supervised models are further handicapped by domain shifts, when the labeled dataset, despite being large enough, fails to cover different protocols or ethnicities. In this paper, we introduce \emph{extreme consistency}, which overcomes the above limitations, by maximally leveraging unlabeled data from the same or a different domain in a teacher-student semi-supervised paradigm. Extreme consistency is the process of sending an extreme transformation of a given image to the student network and then constraining its prediction to be consistent with the teacher network's prediction for the untransformed image. The extreme nature of our consistency loss distinguishes our method from related works that yield suboptimal performance by exercising only mild prediction consistency. Our method is 1) auto-didactic, as it requires no extra expert annotations; 2) versatile, as it handles both domain shift and limited annotation problems; 3) generic, as it is readily applicable to classification, segmentation, and detection tasks; and 4) simple to implement, as it requires no adversarial training. We evaluate our method for the tasks of lesion and retinal vessel segmentation in skin and fundus images. Our experiments demonstrate a significant performance gain over both modern supervised networks and recent semi-supervised models. This performance is attributed to the strong regularization enforced by extreme consistency, which enables the student network to learn how to handle extreme variants of both labeled and unlabeled images. This enhances the network's ability to tackle the inevitable same- and cross-domain data variability during inference.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge