Exploring Embedding Methods in Binary Hyperdimensional Computing: A Case Study for Motor-Imagery based Brain-Computer Interfaces
Paper and Code
Dec 30, 2018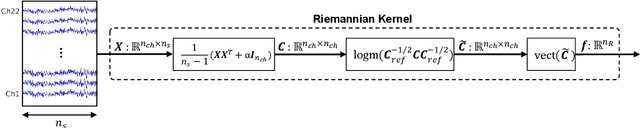
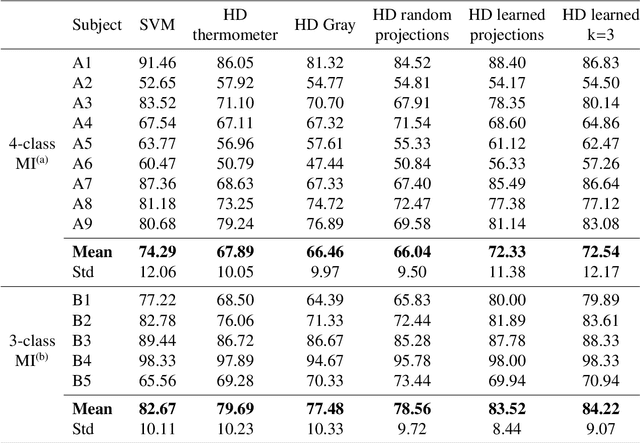
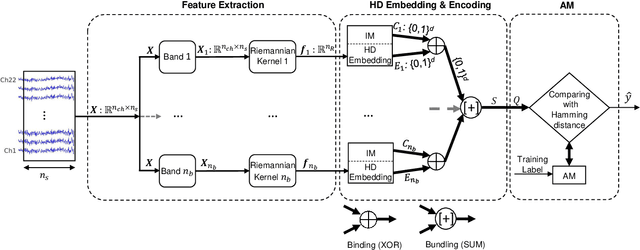
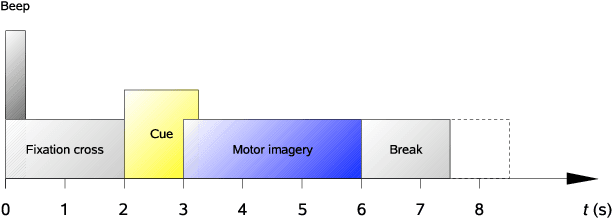
Key properties of brain-inspired hyperdimensional (HD) computing make it a prime candidate for energy-efficient and fast learning in biosignal processing. The main challenge is however to formulate embedding methods that map biosignal measures to a binary HD space. In this paper, we explore variety of such embedding methods and examine them with a challenging application of motor imagery brain-computer interface (MI-BCI) from electroencephalography (EEG) recordings. We explore embedding methods including random projections, quantization based thermometer and Gray coding, and learning HD representations using end-to-end training. All these methods, differing in complexity, aim to represent EEG signals in binary HD space, e.g. with 10,000 bits. This leads to development of a set of HD learning and classification methods that can be selectively chosen (or configured) based on accuracy and/or computational complexity requirements of a given task. We compare them with state-of-the-art linear support vector machine (SVM) on an NVIDIA TX2 board using the 4-class BCI competition IV-2a dataset as well as a new 3-class dataset. Compared to SVM, results on 3-class dataset show that simple thermometer embedding achieves moderate average accuracy (79.56% vs. 82.67%) with 26.8$\times$ faster training time and 22.3$\times$ lower energy; on the other hand, switching to end-to-end training with learned HD representations wipes out these training benefits while boosting the accuracy to 84.22% (1.55% higher than SVM). Similar trend is observed on the 4-class dataset where SVM achieves on average 74.29%: the thermometer embedding achieves 89.9$\times$ faster training time and 58.7$\times$ lower energy, but a lower accuracy (67.09%) than the learned representation of 72.54%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge