Explainable Enrichment-Driven GrAph Reasoner (EDGAR) for Large Knowledge Graphs with Applications in Drug Repurposing
Paper and Code
Sep 27, 2024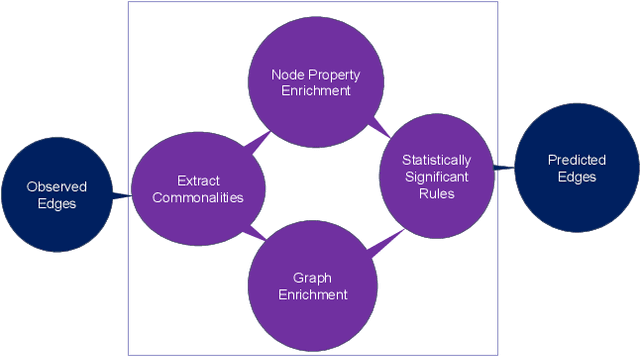
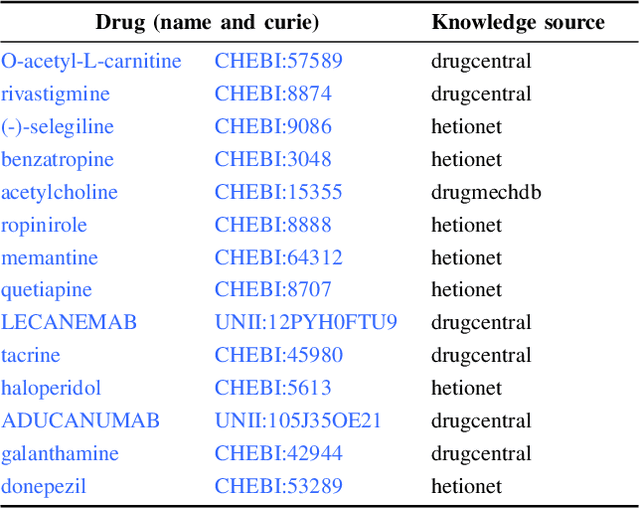
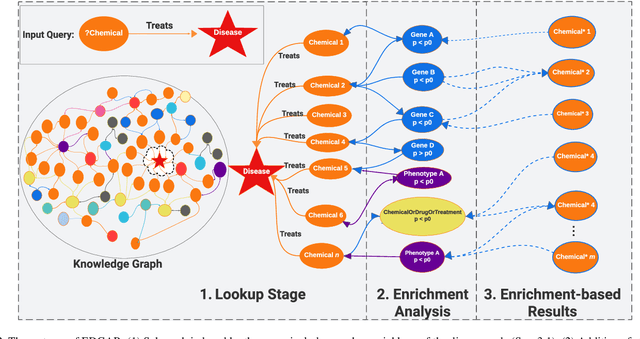
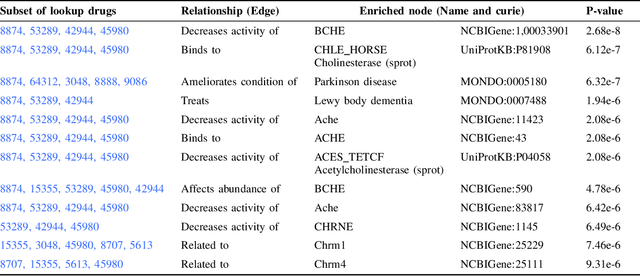
Knowledge graphs (KGs) represent connections and relationships between real-world entities. We propose a link prediction framework for KGs named Enrichment-Driven GrAph Reasoner (EDGAR), which infers new edges by mining entity-local rules. This approach leverages enrichment analysis, a well-established statistical method used to identify mechanisms common to sets of differentially expressed genes. EDGAR's inference results are inherently explainable and rankable, with p-values indicating the statistical significance of each enrichment-based rule. We demonstrate the framework's effectiveness on a large-scale biomedical KG, ROBOKOP, focusing on drug repurposing for Alzheimer disease (AD) as a case study. Initially, we extracted 14 known drugs from the KG and identified 20 contextual biomarkers through enrichment analysis, revealing functional pathways relevant to shared drug efficacy for AD. Subsequently, using the top 1000 enrichment results, our system identified 1246 additional drug candidates for AD treatment. The top 10 candidates were validated using evidence from medical literature. EDGAR is deployed within ROBOKOP, complete with a web user interface. This is the first study to apply enrichment analysis to large graph completion and drug repurposing.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge