Does your model understand genes? A benchmark of gene properties for biological and text models
Paper and Code
Dec 05, 2024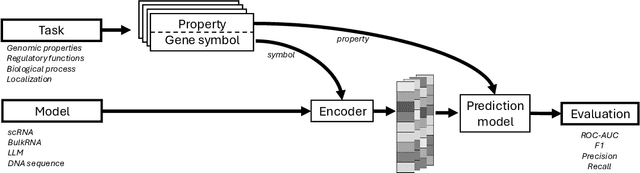
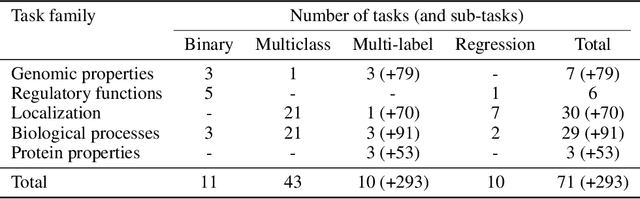
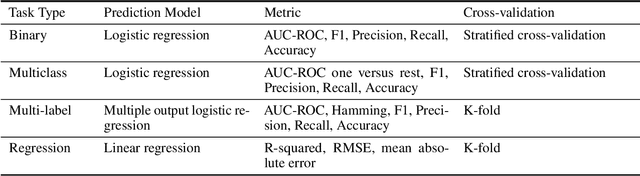

The application of deep learning methods, particularly foundation models, in biological research has surged in recent years. These models can be text-based or trained on underlying biological data, especially omics data of various types. However, comparing the performance of these models consistently has proven to be a challenge due to differences in training data and downstream tasks. To tackle this problem, we developed an architecture-agnostic benchmarking approach that, instead of evaluating the models directly, leverages entity representation vectors from each model and trains simple predictive models for each benchmarking task. This ensures that all types of models are evaluated using the same input and output types. Here we focus on gene properties collected from professionally curated bioinformatics databases. These gene properties are categorized into five major groups: genomic properties, regulatory functions, localization, biological processes, and protein properties. Overall, we define hundreds of tasks based on these databases, which include binary, multi-label, and multi-class classification tasks. We apply these benchmark tasks to evaluate expression-based models, large language models, protein language models, DNA-based models, and traditional baselines. Our findings suggest that text-based models and protein language models generally outperform expression-based models in genomic properties and regulatory functions tasks, whereas expression-based models demonstrate superior performance in localization tasks. These results should aid in the development of more informed artificial intelligence strategies for biological understanding and therapeutic discovery. To ensure the reproducibility and transparency of our findings, we have made the source code and benchmark data publicly accessible for further investigation and expansion at github.com/BiomedSciAI/gene-benchmark.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge