Discrete Morse Sandwich: Fast Computation of Persistence Diagrams for Scalar Data -- An Algorithm and A Benchmark
Paper and Code
Jun 27, 2022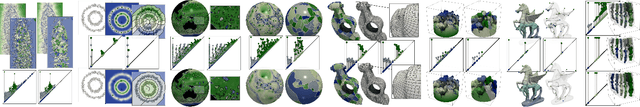
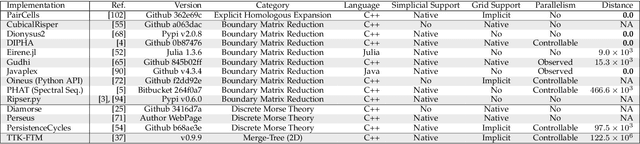
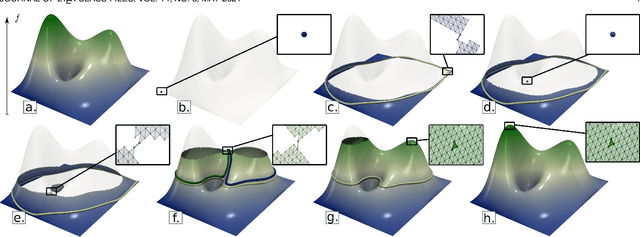
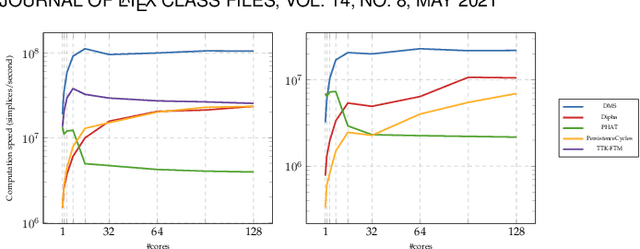
This paper introduces an efficient algorithm for persistence diagram computation, given an input piecewise linear scalar field f defined on a d-dimensional simplicial complex K, with $d \leq 3$. Our method extends the seminal "PairCells" algorithm by introducing three main accelerations. First, we express this algorithm within the setting of discrete Morse theory, which considerably reduces the number of input simplices to consider. Second, we introduce a stratification approach to the problem, that we call "sandwiching". Specifically, minima-saddle persistence pairs ($D_0(f)$) and saddle-maximum persistence pairs ($D_{d-1}(f)$) are efficiently computed by respectively processing with a Union-Find the unstable sets of 1-saddles and the stable sets of (d-1)-saddles. This fast processing of the dimensions 0 and (d-1) further reduces, and drastically, the number of critical simplices to consider for the computation of $D_1(f)$, the intermediate layer of the sandwich. Third, we document several performance improvements via shared-memory parallelism. We provide an open-source implementation of our algorithm for reproducibility purposes. We also contribute a reproducible benchmark package, which exploits three-dimensional data from a public repository and compares our algorithm to a variety of publicly available implementations. Extensive experiments indicate that our algorithm improves by two orders of magnitude the time performance of the seminal "PairCells" algorithm it extends. Moreover, it also improves memory footprint and time performance over a selection of 14 competing approaches, with a substantial gain over the fastest available approaches, while producing a strictly identical output. We illustrate the utility of our contributions with an application to the fast and robust extraction of persistent 1-dimensional generators on surfaces, volume data and high-dimensional point clouds.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge