Directional Message Passing on Molecular Graphs via Synthetic Coordinates
Paper and Code
Nov 08, 2021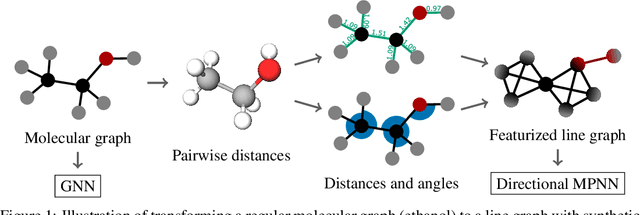
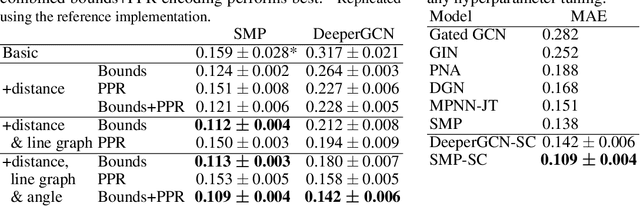

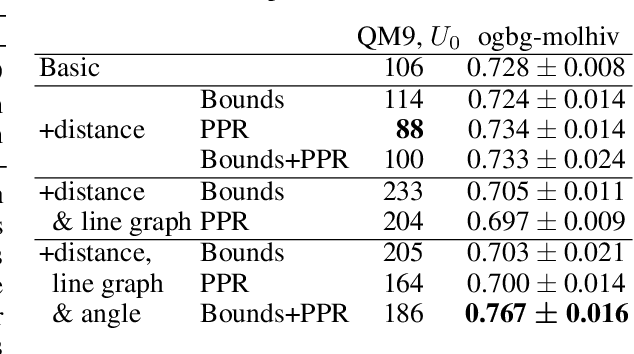
Graph neural networks that leverage coordinates via directional message passing have recently set the state of the art on multiple molecular property prediction tasks. However, they rely on atom position information that is often unavailable, and obtaining it is usually prohibitively expensive or even impossible. In this paper we propose synthetic coordinates that enable the use of advanced GNNs without requiring the true molecular configuration. We propose two distances as synthetic coordinates: Distance bounds that specify the rough range of molecular configurations, and graph-based distances using a symmetric variant of personalized PageRank. To leverage both distance and angular information we propose a method of transforming normal graph neural networks into directional MPNNs. We show that with this transformation we can reduce the error of a normal graph neural network by 55% on the ZINC benchmark. We furthermore set the state of the art on ZINC and coordinate-free QM9 by incorporating synthetic coordinates in the SMP and DimeNet++ models. Our implementation is available online.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge