Deep Learning for Plant Identification and Disease Classification from Leaf Images: Multi-prediction Approaches
Paper and Code
Oct 25, 2023
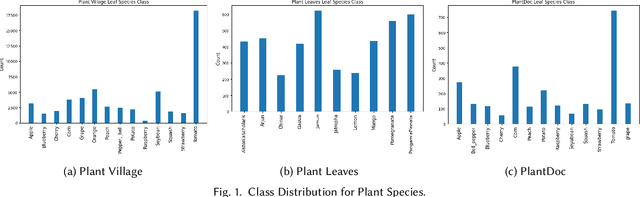
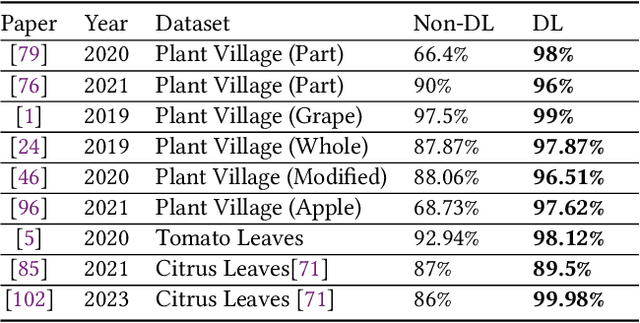
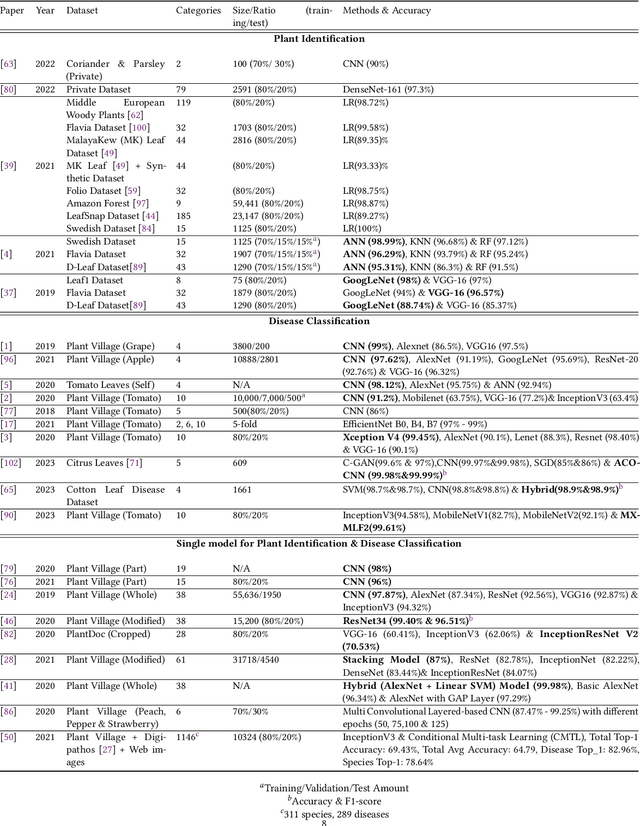
Deep learning plays an important role in modern agriculture, especially in plant pathology using leaf images where convolutional neural networks (CNN) are attracting a lot of attention. While numerous reviews have explored the applications of deep learning within this research domain, there remains a notable absence of an empirical study to offer insightful comparisons due to the employment of varied datasets in the evaluation. Furthermore, a majority of these approaches tend to address the problem as a singular prediction task, overlooking the multifaceted nature of predicting various aspects of plant species and disease types. Lastly, there is an evident need for a more profound consideration of the semantic relationships that underlie plant species and disease types. In this paper, we start our study by surveying current deep learning approaches for plant identification and disease classification. We categorise the approaches into multi-model, multi-label, multi-output, and multi-task, in which different backbone CNNs can be employed. Furthermore, based on the survey of existing approaches in plant pathology and the study of available approaches in machine learning, we propose a new model named Generalised Stacking Multi-output CNN (GSMo-CNN). To investigate the effectiveness of different backbone CNNs and learning approaches, we conduct an intensive experiment on three benchmark datasets Plant Village, Plant Leaves, and PlantDoc. The experimental results demonstrate that InceptionV3 can be a good choice for a backbone CNN as its performance is better than AlexNet, VGG16, ResNet101, EfficientNet, MobileNet, and a custom CNN developed by us. Interestingly, empirical results support the hypothesis that using a single model can be comparable or better than using two models. Finally, we show that the proposed GSMo-CNN achieves state-of-the-art performance on three benchmark datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge