Deep Generative Endmember Modeling: An Application to Unsupervised Spectral Unmixing
Paper and Code
Feb 14, 2019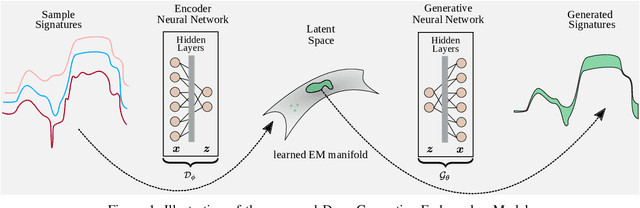
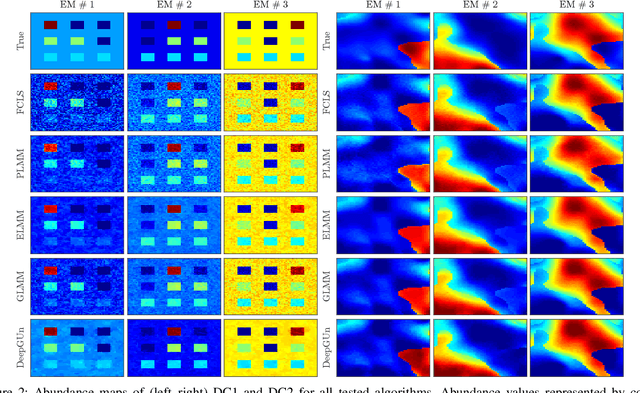
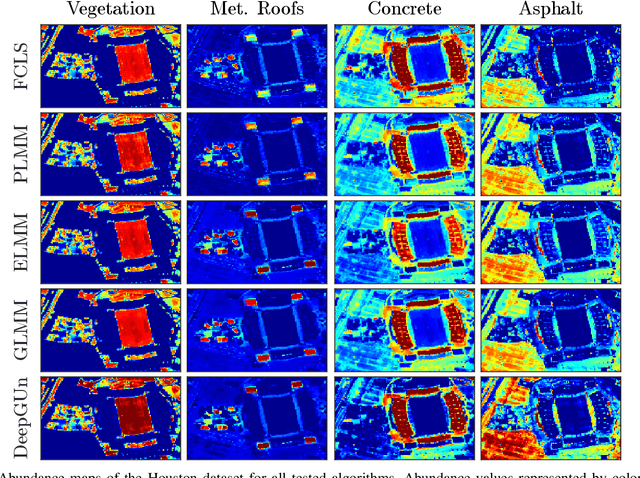
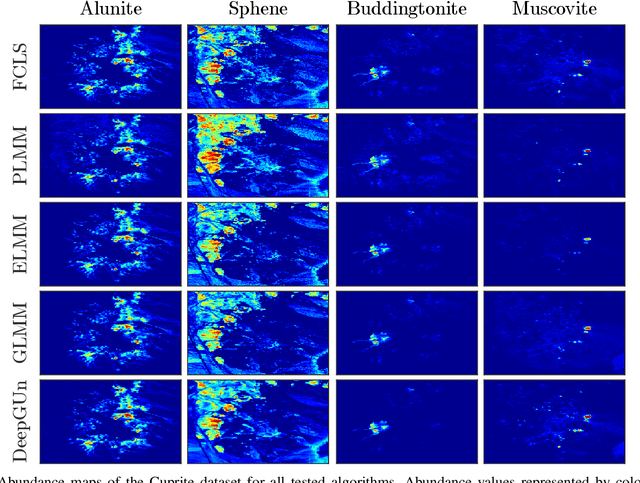
Endmember (EM) spectral variability can greatly impact the performance of standard hyperspectral image analysis algorithms. Extended parametric models have been successfully applied to account for the EM spectral variability. However, these models still lack the compromise between flexibility and low-dimensional representation that is necessary to properly explore the fact that spectral variability is often confined to a low-dimensional manifold in real scenes. In this paper we propose to learn a spectral variability model directly form the observed data, instead of imposing it \emph{a priori}. This is achieved through a deep generative EM model, which is estimated using a variational autoencoder (VAE). The encoder and decoder that compose the generative model are trained using pure pixel information extracted directly from the observed image, what allows for an unsupervised formulation. The proposed EM model is applied to the solution of a spectral unmixing problem, which we cast as an alternating nonlinear least-squares problem that is solved iteratively with respect to the abundances and to the low-dimensional representations of the EMs in the latent space of the deep generative model. Simulations using both synthetic and real data indicate that the proposed strategy can outperform the competing state-of-the-art algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge