DDIPrompt: Drug-Drug Interaction Event Prediction based on Graph Prompt Learning
Paper and Code
Feb 18, 2024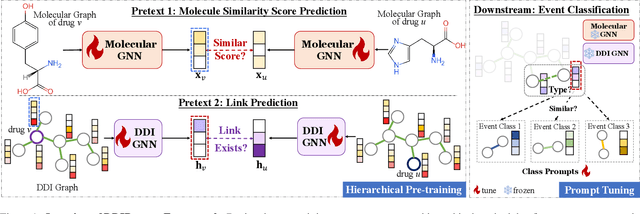
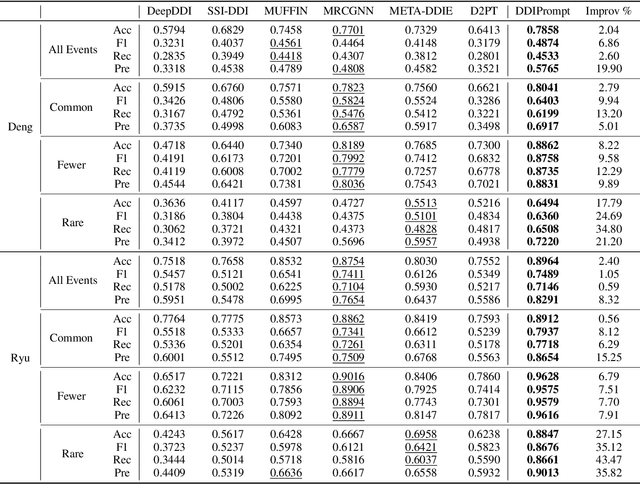
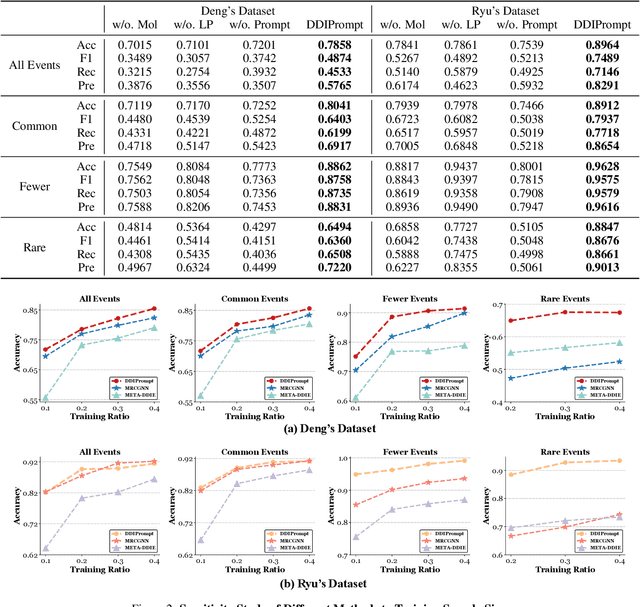
Recently, Graph Neural Networks have become increasingly prevalent in predicting adverse drug-drug interactions (DDI) due to their proficiency in modeling the intricate associations between atoms and functional groups within and across drug molecules. However, they are still hindered by two significant challenges: (1) the issue of highly imbalanced event distribution, which is a common but critical problem in medical datasets where certain interactions are vastly underrepresented. This imbalance poses a substantial barrier to achieving accurate and reliable DDI predictions. (2) the scarcity of labeled data for rare events, which is a pervasive issue in the medical field where rare yet potentially critical interactions are often overlooked or under-studied due to limited available data. In response, we offer DDIPrompt, an innovative panacea inspired by the recent advancements in graph prompting. Our framework aims to address these issues by leveraging the intrinsic knowledge from pre-trained models, which can be efficiently deployed with minimal downstream data. Specifically, to solve the first challenge, DDIPrompt employs augmented links between drugs, considering both structural and interactive proximity. It features a hierarchical pre-training strategy that comprehends intra-molecular structures and inter-molecular interactions, fostering a comprehensive and unbiased understanding of drug properties. For the second challenge, we implement a prototype-enhanced prompting mechanism during inference. This mechanism, refined by few-shot examples from each category, effectively harnesses the rich pre-training knowledge to enhance prediction accuracy, particularly for these rare but crucial interactions. Comprehensive evaluations on two benchmark datasets demonstrate the superiority of DDIPrompt, particularly in predicting rare DDI events.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge