Coordinating Cross-modal Distillation for Molecular Property Prediction
Paper and Code
Nov 30, 2022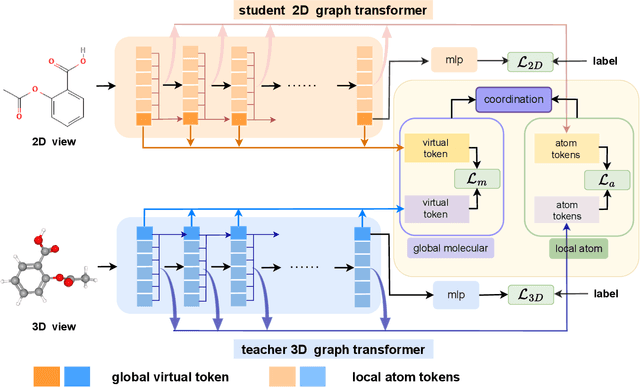
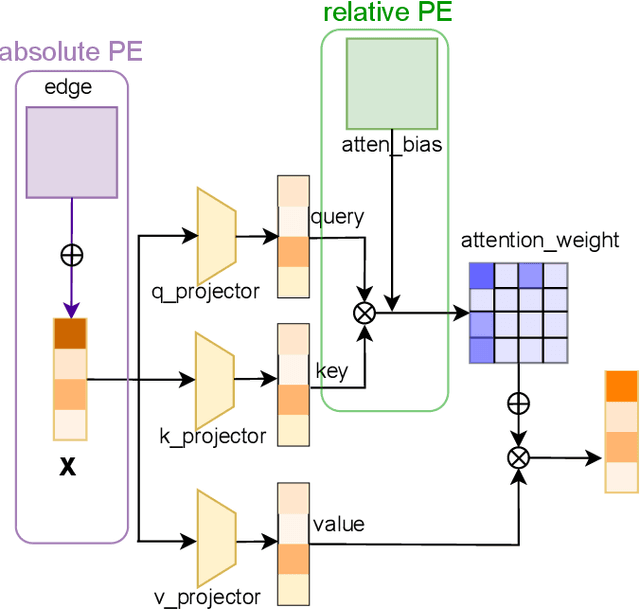
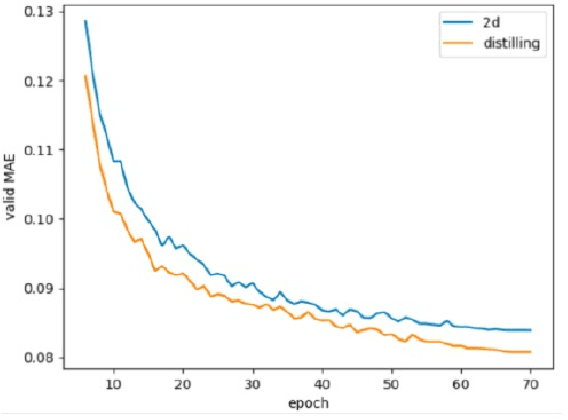
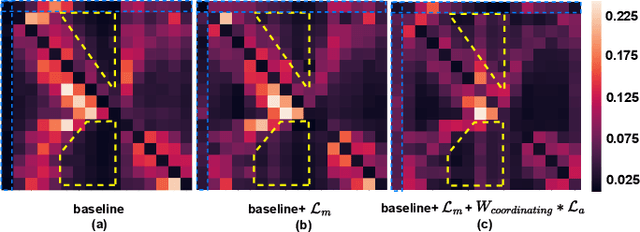
In recent years, molecular graph representation learning (GRL) has drawn much more attention in molecular property prediction (MPP) problems. The existing graph methods have demonstrated that 3D geometric information is significant for better performance in MPP. However, accurate 3D structures are often costly and time-consuming to obtain, limiting the large-scale application of GRL. It is an intuitive solution to train with 3D to 2D knowledge distillation and predict with only 2D inputs. But some challenging problems remain open for 3D to 2D distillation. One is that the 3D view is quite distinct from the 2D view, and the other is that the gradient magnitudes of atoms in distillation are discrepant and unstable due to the variable molecular size. To address these challenging problems, we exclusively propose a distillation framework that contains global molecular distillation and local atom distillation. We also provide a theoretical insight to justify how to coordinate atom and molecular information, which tackles the drawback of variable molecular size for atom information distillation. Experimental results on two popular molecular datasets demonstrate that our proposed model achieves superior performance over other methods. Specifically, on the largest MPP dataset PCQM4Mv2 served as an "ImageNet Large Scale Visual Recognition Challenge" in the field of graph ML, the proposed method achieved a 6.9% improvement compared with the best works. And we obtained fourth place with the MAE of 0.0734 on the test-challenge set for OGB-LSC 2022 Graph Regression Task. We will release the code soon.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge