Cometh: A continuous-time discrete-state graph diffusion model
Paper and Code
Jun 10, 2024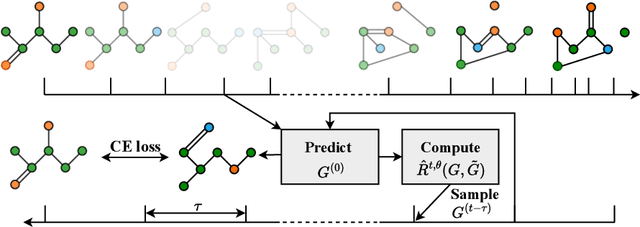
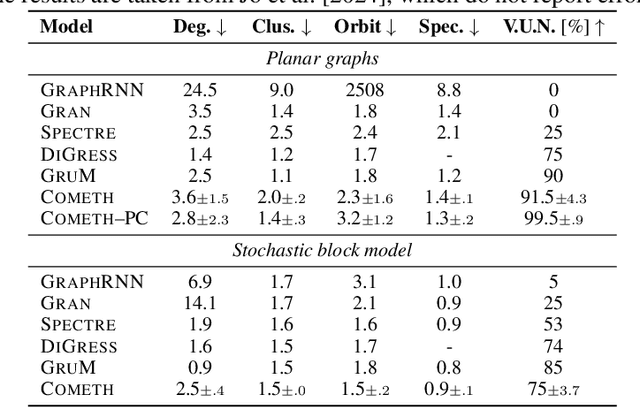
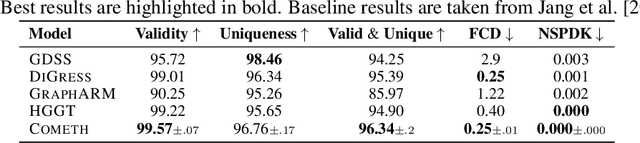
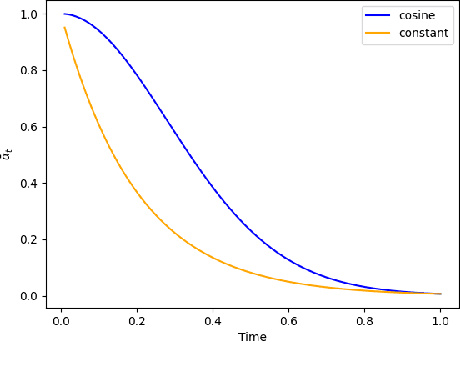
Discrete-state denoising diffusion models led to state-of-the-art performance in graph generation, especially in the molecular domain. Recently, they have been transposed to continuous time, allowing more flexibility in the reverse process and a better trade-off between sampling efficiency and quality. Here, to leverage the benefits of both approaches, we propose Cometh, a continuous-time discrete-state graph diffusion model, integrating graph data into a continuous-time diffusion model framework. Empirically, we show that integrating continuous time leads to significant improvements across various metrics over state-of-the-art discrete-state diffusion models on a large set of molecular and non-molecular benchmark datasets.
* 23 pages
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge