Classification on Large Networks: A Quantitative Bound via Motifs and Graphons
Paper and Code
Oct 24, 2017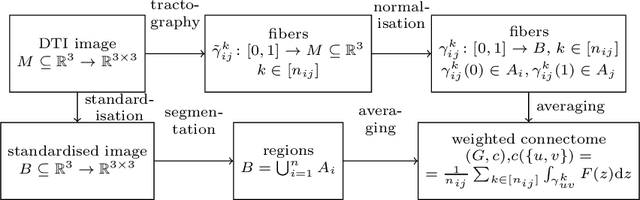
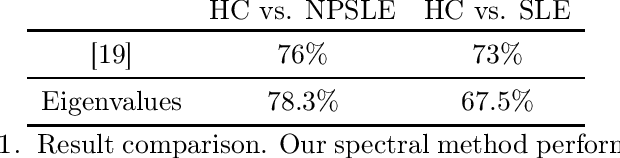
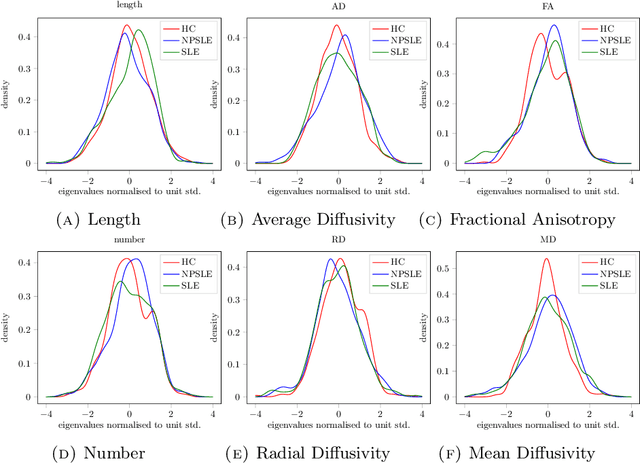
When each data point is a large graph, graph statistics such as densities of certain subgraphs (motifs) can be used as feature vectors for machine learning. While intuitive, motif counts are expensive to compute and difficult to work with theoretically. Via graphon theory, we give an explicit quantitative bound for the ability of motif homomorphisms to distinguish large networks under both generative and sampling noise. Furthermore, we give similar bounds for the graph spectrum and connect it to homomorphism densities of cycles. This results in an easily computable classifier on graph data with theoretical performance guarantee. Our method yields competitive results on classification tasks for the autoimmune disease Lupus Erythematosus.
* 17 pages, 2 figures, 1 table
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge