Anomaly Detection in Retinal Images using Multi-Scale Deep Feature Sparse Coding
Paper and Code
Jan 27, 2022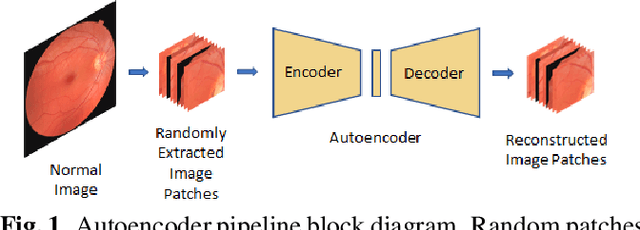
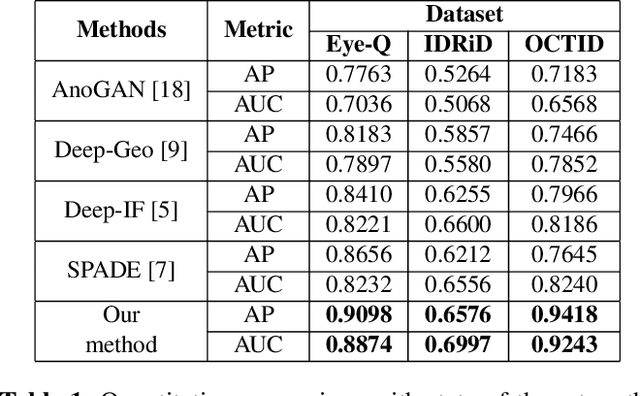
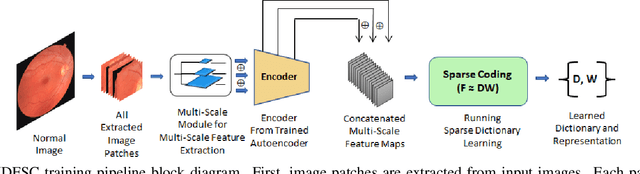
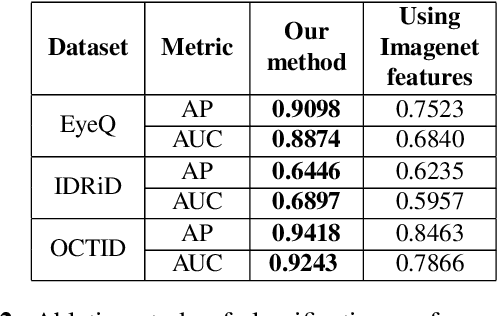
Convolutional Neural Network models have successfully detected retinal illness from optical coherence tomography (OCT) and fundus images. These CNN models frequently rely on vast amounts of labeled data for training, difficult to obtain, especially for rare diseases. Furthermore, a deep learning system trained on a data set with only one or a few diseases cannot detect other diseases, limiting the system's practical use in disease identification. We have introduced an unsupervised approach for detecting anomalies in retinal images to overcome this issue. We have proposed a simple, memory efficient, easy to train method which followed a multi-step training technique that incorporated autoencoder training and Multi-Scale Deep Feature Sparse Coding (MDFSC), an extended version of normal sparse coding, to accommodate diverse types of retinal datasets. We achieve relative AUC score improvement of 7.8\%, 6.7\% and 12.1\% over state-of-the-art SPADE on Eye-Q, IDRiD and OCTID datasets respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge