Anfinsen Goes Neural: a Graphical Model for Conditional Antibody Design
Paper and Code
Feb 08, 2024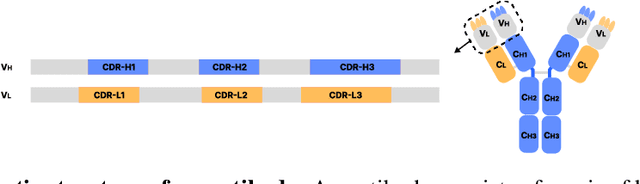
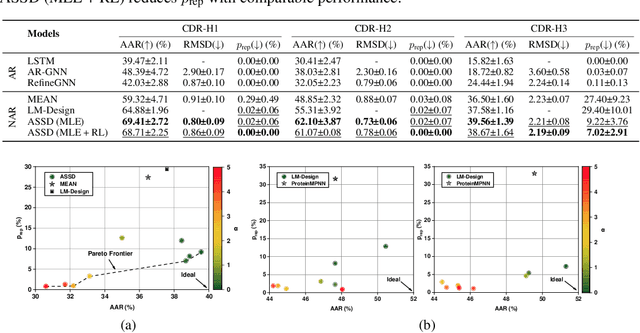
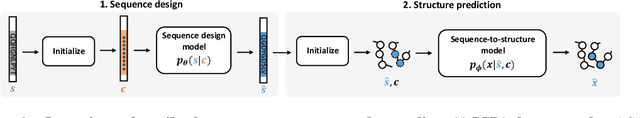
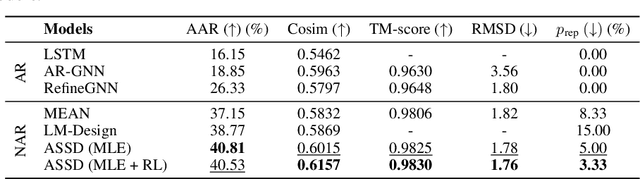
Antibody design plays a pivotal role in advancing therapeutics. Although deep learning has made rapid progress in this field, existing methods make limited use of general protein knowledge and assume a graphical model (GM) that violates empirical findings on proteins. To address these limitations, we present Anfinsen Goes Neural (AGN), a graphical model that uses a pre-trained protein language model (pLM) and encodes a seminal finding on proteins called Anfinsen's dogma. Our framework follows a two-step process of sequence generation with pLM and structure prediction with graph neural network (GNN). Experiments show that our approach outperforms state-of-the-art results on benchmark experiments. We also address a critical limitation of non-autoregressive models -- namely, that they tend to generate unrealistic sequences with overly repeating tokens. To resolve this, we introduce a composition-based regularization term to the cross-entropy objective that allows an efficient trade-off between high performance and low token repetition. We demonstrate that our approach establishes a Pareto frontier over the current state-of-the-art. Our code is available at https://github.com/lkny123/AGN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge