Adaptive Residue-wise Profile Fusion for Low Homologous Protein SecondaryStructure Prediction Using External Knowledge
Paper and Code
Aug 05, 2021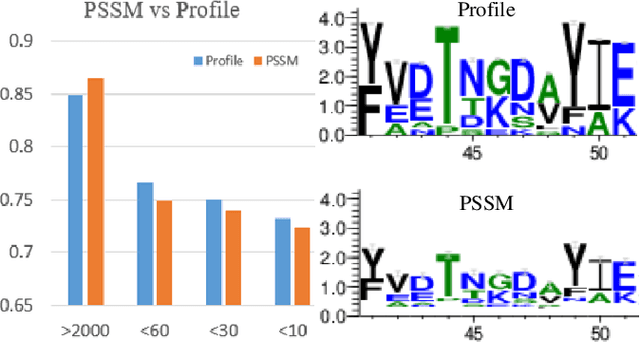
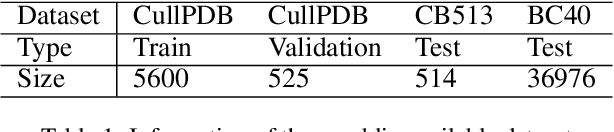
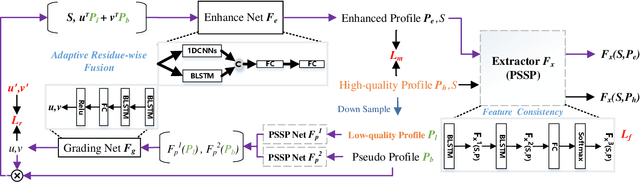
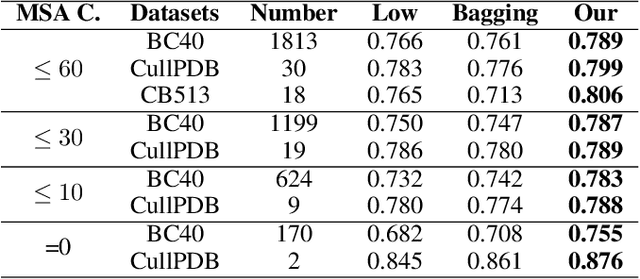
Protein secondary structure prediction (PSSP) is essential for protein function analysis. However, for low homologous proteins, the PSSP suffers from insufficient input features. In this paper, we explicitly import external self-supervised knowledge for low homologous PSSP under the guidance of residue-wise profile fusion. In practice, we firstly demonstrate the superiority of profile over Position-Specific Scoring Matrix (PSSM) for low homologous PSSP. Based on this observation, we introduce the novel self-supervised BERT features as the pseudo profile, which implicitly involves the residue distribution in all native discovered sequences as the complementary features. Further-more, a novel residue-wise attention is specially designed to adaptively fuse different features (i.e.,original low-quality profile, BERT based pseudo profile), which not only takes full advantage of each feature but also avoids noise disturbance. Be-sides, the feature consistency loss is proposed to accelerate the model learning from multiple semantic levels. Extensive experiments confirm that our method outperforms state-of-the-arts (i.e.,4.7%forextremely low homologous cases on BC40 dataset).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge