A Translate-Edit Model for Natural Language Question to SQL Query Generation on Multi-relational Healthcare Data
Paper and Code
Jul 28, 2019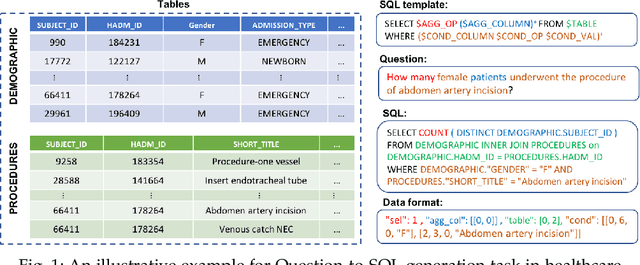
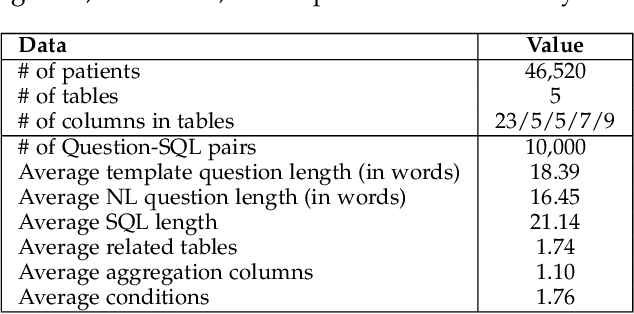
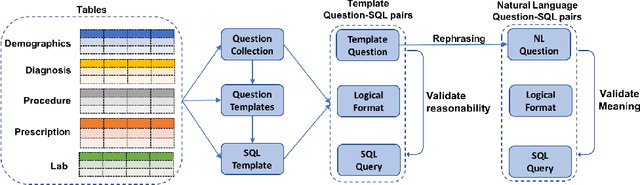
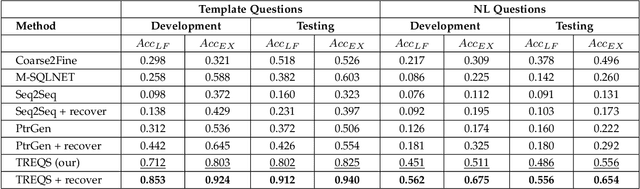
Electronic health record (EHR) data contains most of the important patient health information and is typically stored in a relational database with multiple tables. One important way for doctors to make use of EHR data is to retrieve intuitive information by posing a sequence of questions against it. However, due to a large amount of information stored in it, effectively retrieving patient information from EHR data in a short time is still a challenging issue for medical experts since it requires a good understanding of a query language to get access to the database. We tackle this challenge by developing a deep learning based approach that can translate a natural language question on multi-relational EHR data into its corresponding SQL query, which is referred to as a Question-to-SQL generation task. Most of the existing methods cannot solve this problem since they primarily focus on tackling the questions related to a single table under the table-aware assumption. While in our problem, it is possible that questions asked by clinicians are related to multiple unspecified tables. In this paper, we first create a new question to query dataset designed for healthcare to perform the Question-to-SQL generation task, named MIMICSQL, based on a publicly available electronic medical database. To address the challenge of generating queries on multi-relational databases from natural language questions, we propose a TRanslate-Edit Model for Question-to-SQL query (TREQS), which adopts the sequence-to-sequence model to directly generate SQL query for a given question, and further edits it with an attentive-copying mechanism and task-specific look-up tables. Both quantitative and qualitative experimental results indicate the flexibility and efficiency of our proposed method in tackling challenges that are unique in MIMICSQL.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge