A Survey On 3D Inner Structure Prediction from its Outer Shape
Paper and Code
Feb 11, 2020
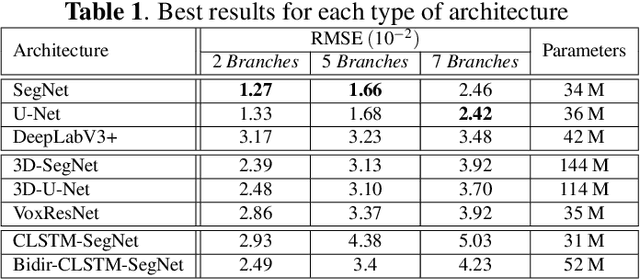
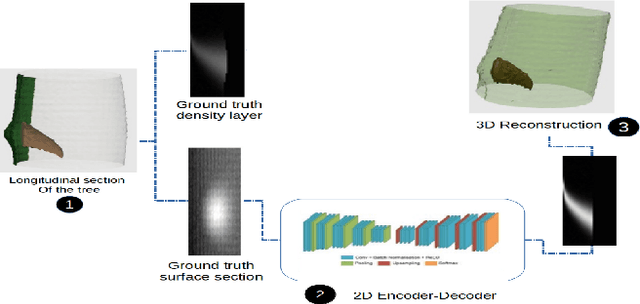
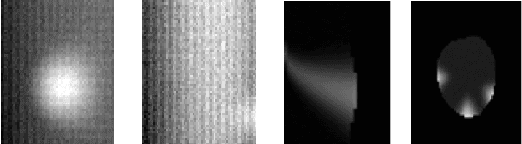
The analysis of the internal structure of trees is highly important for both forest experts, biological scientists, and the wood industry. Traditionally, CT-scanners are considered as the most efficient way to get an accurate inner representation of the tree. However, this method requires an important investment and reduces the cost-effectiveness of this operation. Our goal is to design neural-network-based methods to predict the internal density of the tree from its external bark shape. This paper compares different image-to-image(2D), volume-to-volume(3D) and Convolutional Long Short Term Memory based neural network architectures in the context of the prediction of the defect distribution inside trees from their external bark shape. Those models are trained on a synthetic dataset of 1800 CT-scanned look-like volumetric structures of the internal density of the trees and their corresponding external surface.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge