A step towards neural genome assembly
Paper and Code
Nov 10, 2020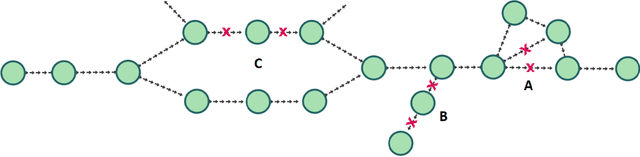
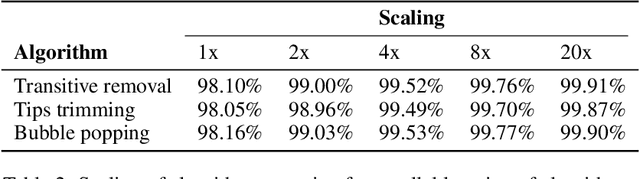
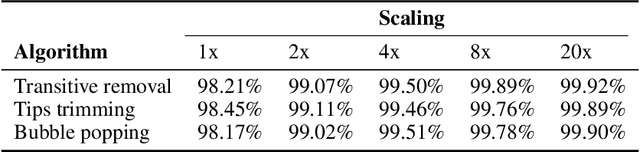

De novo genome assembly focuses on finding connections between a vast amount of short sequences in order to reconstruct the original genome. The central problem of genome assembly could be described as finding a Hamiltonian path through a large directed graph with a constraint that an unknown number of nodes and edges should be avoided. However, due to local structures in the graph and biological features, the problem can be reduced to graph simplification, which includes removal of redundant information. Motivated by recent advancements in graph representation learning and neural execution of algorithms, in this work we train the MPNN model with max-aggregator to execute several algorithms for graph simplification. We show that the algorithms were learned successfully and can be scaled to graphs of sizes up to 20 times larger than the ones used in training. We also test on graphs obtained from real-world genomic data---that of a lambda phage and E. coli.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge