A Novel Learning Algorithm for Bayesian Network and Its Efficient Implementation on GPU
Paper and Code
Oct 18, 2012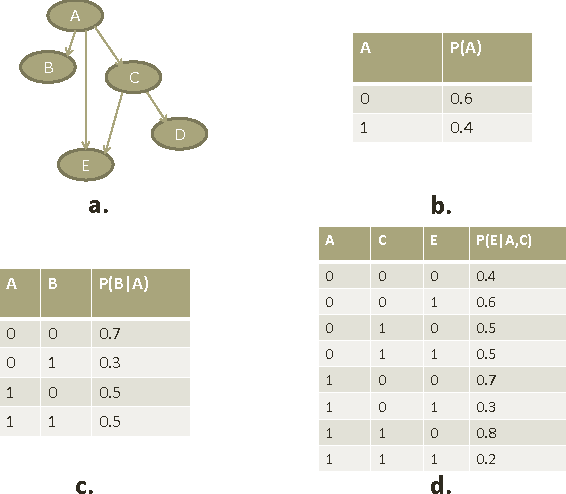
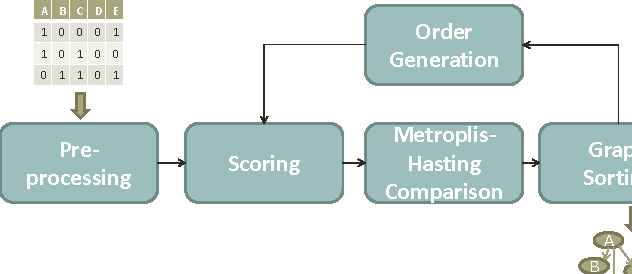
Computational inference of causal relationships underlying complex networks, such as gene-regulatory pathways, is NP-complete due to its combinatorial nature when permuting all possible interactions. Markov chain Monte Carlo (MCMC) has been introduced to sample only part of the combinations while still guaranteeing convergence and traversability, which therefore becomes widely used. However, MCMC is not able to perform efficiently enough for networks that have more than 15~20 nodes because of the computational complexity. In this paper, we use general purpose processor (GPP) and general purpose graphics processing unit (GPGPU) to implement and accelerate a novel Bayesian network learning algorithm. With a hash-table-based memory-saving strategy and a novel task assigning strategy, we achieve a 10-fold acceleration per iteration than using a serial GPP. Specially, we use a greedy method to search for the best graph from a given order. We incorporate a prior component in the current scoring function, which further facilitates the searching. Overall, we are able to apply this system to networks with more than 60 nodes, allowing inferences and modeling of bigger and more complex networks than current methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge